With highly sensitive single cell RNA-seq (scRNAseq), our study highlights the effects of blood storage on PBMCs, e.g. gene sets highly relevant to human diseases (NF-kB, AP-1 signaling) are upregulated, and advocates for scRNAseq/bulk RNAseq experiments of PBMCs from fresh blood.
- AP-1
- NF-kB
- peripheral blood mononuclear cells
- single cell RNA-seq
- transcriptome
1. Introduction
Introduction
Peripheral blood mononuclear cells (PBMCs) are widely used as a model in the study of different human diseases. There is often a time delay from blood collection to PBMC isolation during the sampling process, which can result in an experimental bias, particularly when performing highly sensitive single cell RNA-seq (scRNAseq) studies. This study examined the impact of different time periods from blood draw to PBMC isolation on the subsequent transcriptome profiling of different cell types in PBMCs by scRNAseq using the 10X Chromium Single Cell Gene Expression assay. scRNAseq in this study was done using 10X Chromium Single Cell 3′ Gene Expression Solution (10x Genomics, v3 chemistry).
2. New discoveries
New discoveries
Examining the five major cell types constituting the PBMC cell population, i.e., CD4+ T cells, CD8+ T cells, NK cells, monocytes, and B cells, both common changes and cell-type-specific changes were observed in the single cell transcriptome profiling over time. In particular, the upregulation of genes regulated by NF-kB in response to TNF was observed in all five cell types. Significant changes in key genes involved in AP-1 signaling were also observed. RBC contamination was a major issue in stored blood, whereas RBC adherence had no direct impact on the cell transcriptome. The expression levels and statistics of individual genes in each cell type are shown in the Supplementary Data 1–5 of this article.
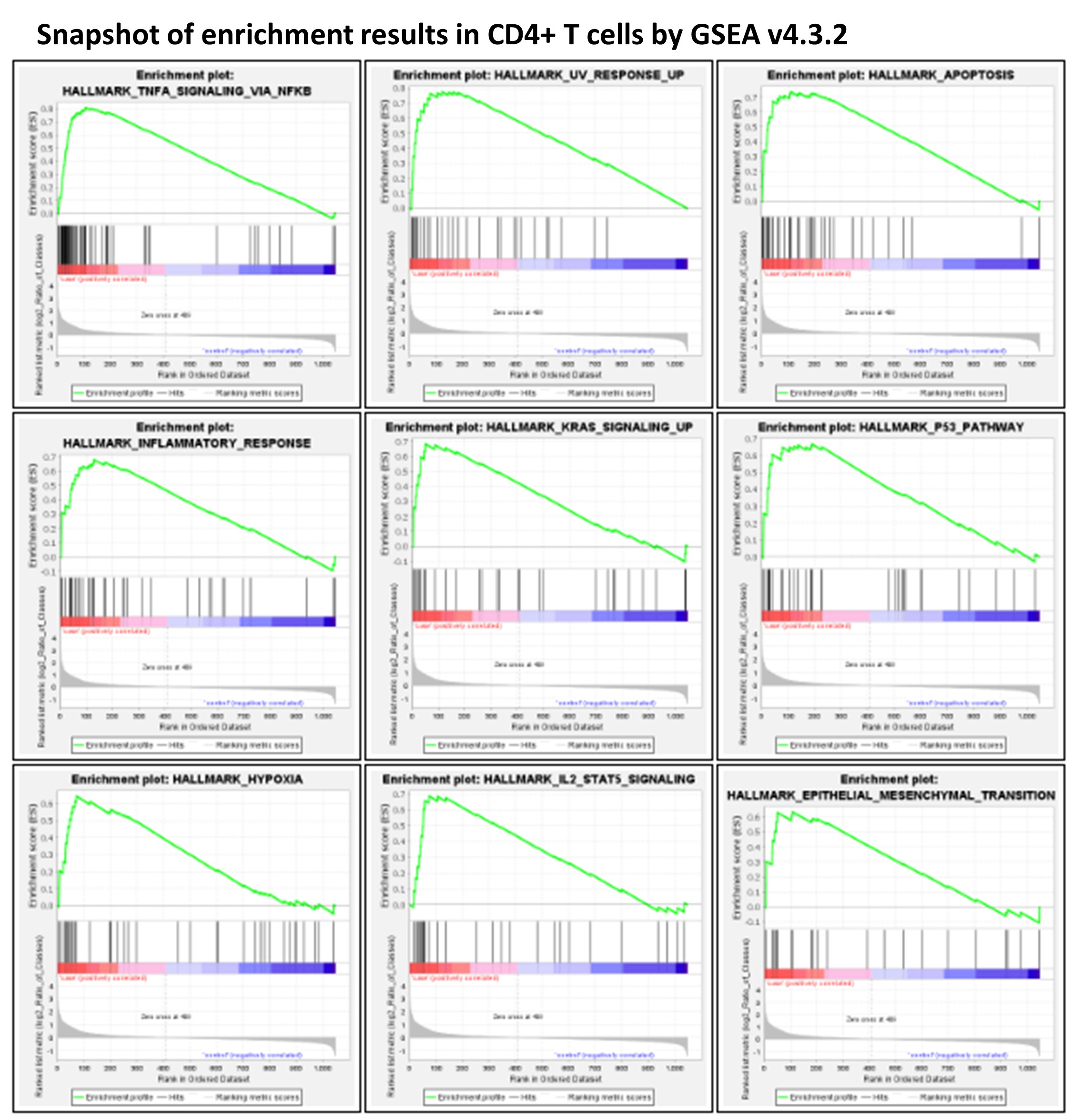
- GSEA Analysis
GSEA analysis highlighted that a number of MSigDB Hallmark gene sets(Liberzon, Birger et al. 2015) were significantly impacted in stored blood (FDR < 0.1). In particular, the HALLMARK_TNFA_SIGNALING_VIA_NFKB gene set was upregulated in all five cell types; the HALLMARK_APOPTOSIS gene set was upregulated in CD4+ T cells and monocyteplications. In contrast, expression changes of the gene sets HALLMARK_MYC_TARGETS_V1 and HALLMARK_OXIDATIVE_PHOSPHORYLATION showed heterogeneity across different cell types. HALLMARK_MYC_TARGETS_V1 and HALLMARK_OXIDATIVE_PHOSPHORYLATION were downregulated with statistical significance in CD4+ T cells, while HALLMARK_MYC_TARGETS_V1 had positive ES scores in CD8+ T cells and B cells, and HALLMARK_OXIDATIVE_PHOSPHORYLATION had a positive ES in monocytes.
According to previous studies, both granulocyte activation(Kataranovski, Kataranovski et al. 1998, McKenna, Beatty et al. 2009) and RBC contamination(Song, Goodwin et al. 2002) due to blood storage might cause upregulation of the HALLMARK_TNFA_SIGNALING_VIA_NFKB gene set, i.e., genes regulated by nuclear factor kappa B (NF-κB) in response to tumor necrosis factor (TNF). Granulocyte activation is correlated with activated TNFα signaling in different cell types(Presicce, Cappelletti et al. 2020), while all five cell types in our study that had upregulated HALLMARK_TNFA_SIGNALING_VIA_NFKB.
- Specific DE Genes by Blood Storage
Indhis study hividual DE genes with at least 1.5 fold change were examined. Among 24,800 genes in the scRNAseq assay, the DE genes in stored blood are significantly replicable in the two individuals, for both upregulated genes and downregulated genes in each cell type (with high statistical significance by Chi-square test). The significant replicability demonstrated the validity of the observed effects hlights the effects of blood storage on mRNA levels. As a powerful tool for transcriptome profiling at a single cell level, the scRNAseq assay demonstrated statistical power to identify DE genes by comparing multiple cells from each cell type in each sample. The statistical significances of the DE genes were adjusted for multiple comparisons.
- CD4+ T Cells and RBC Adherence
With the contamination of RBCs as an issue in stored blood, it is important to investigate whether the above observed changes in transcriptome in PBMCs were related to the effects of RBC adherence on PBMCs. For this purpose, we investigated the effects of RBC adherence in CD4+ T cells by comparing the transcriptomes of CD4+ T cells with (log2 value of CD4 > 1 and log2 value of HBA1 > 1) vs. without (log2 value of CD4 > 1 and log2 value of HBA1 ≤ 1) the HBA1 feature (Supplementary Data 6 of this article). GSEA analysis showed no significant Hallmark gene sets. Interestingly, the two upregulated gene sets, HALLMARK_TNFA_SIGNALING_VIA_NFKB and HALLMARK_APOPTOSIS, in PBMCs from stored blood have negative ES scores (i.e., no up-regulation) in CD4+ T cells with positive HBA1, implying that the upregulation of these gene sets in stored blood was not due to RBC adherence. Except for the three hemoglobin genes, including HBA1, hemoglobin subunit alpha 2 (HBA2), and hemoglobin subunit beta (HBB), no other gene showed statistical significance when comparing the transcriptomes of CD4+ T cells with vs. without HBA1 features.
Although a previous study suggested that RBC contamination might increase TNF expression by PBMCs(Song, Goodwin et al. 2002), our study showed no direct effects of RBC adherence on the transcriptomes of CD4+ T cells.
3. Applications
This study highlights the effects of blood storage on PBMCs. Some mame major changes of transcriptome highly relevant to human diseases (e.g. NF-kB, AP-1 signaling) in stored blood as shown by this study may not be corrected by computational approaches. With the scRNAseq technology as a highly sensitive and powerful tool, PBMCs extracted from fresh blood are needed for performing transcriptome studies, especially studies with a case–control design.
It is a common scenario that in a study with a case-control design, the patient samples and the healthy controls were collected separately. According to this study, this may cause experimental bias related to blood storage. There are two solutions that may help: (1) to isolate PBMCs from fresh blood for both cases and controls; (2) to adopt a pair-wise experimental design, i.e. to collect blood samples and isolate PBMCs by case/control pairs.
For [1][2][3][4][5].more details, please see https://www.mdpi.com/2073-4425/14/1/142