Polyphenols are valuable natural antioxidants present in diet that likely mitigate aging effects, neurodegenerative conditions, and other diseases. However, because of their poor absorption in the gut and consequent low concentration in biological fluids (µM range), reservations about polyphenol antioxidant efficiency have been raised. The π-π interaction between one polyphenol ring and superoxide is associated with oxidation of the latter due to transfer of its unpaired electron to a polyphenolic aromatic ring, and consequent formation of a molecule of O2 (one product of SOD action). Mechanistically, it is very difficult to establish if this π-π interaction proceeds before or after the most common mode of scavenging superoxide, e.g., abstraction of an aromatic polyphenol H(hydroxyl), which then is used to form H2O2 (the other molecule produced by SOD action). At the end of this cycle of superoxide scavenging, 4-methyl-7,8-di-hydroxy-coumarin and the flavonoid galangin reform themselves. An alternative mechanistic pathway by galangin forms the η-(H2O2)-galangin-η-O2 complex that includes additional H2O2 and O2 molecules. Another mode of action is seen with the chalcone butein, in which the polyphenol system incorporates a molecule of O2, e.g., a η-O2-butein complex is formed, ready for additional scavenging.
- superoxide dismutase
- polyphenol
- superoxide
- antioxidant
1. Introduction
2. 4-Methyl-7,8-di-hydroxy Coumarin
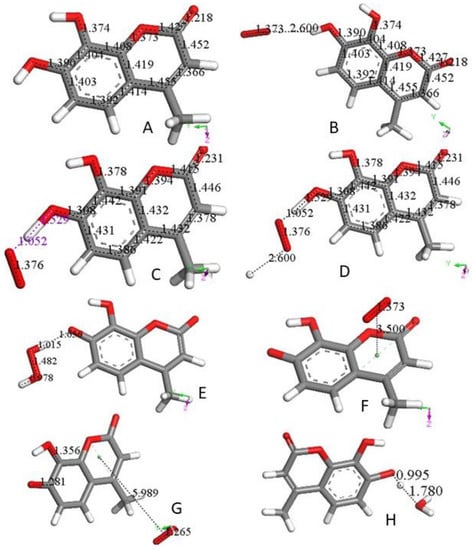
4-Methyl-7,8-di-hydroxy coumarin, as shown in Figure 1A, is an antioxidant [11,12], which can be associated with a standard superoxide scavenging mechanism through polyphenol hydrogen atom abstraction. Computational results describe the interaction between one polyphenol H(hydroxyl) and the radical; Figure 1B shows the initial van der Waals interaction between the two species, which we call type σ. Upon DFT geometry optimization, this system evolves toward formation of an HO2- species, separated 1.529 Å from the remaining radical semiquinone, having the unpaired electron located in the reacted ring, Figure 1C. Next, the Figure 1C DFT minimized structure has HO2− anion posed at van der Waals distance to a proton (2.60 Å) in Figure 1D. Finally, upon geometry minimization, this evolves toward H2O2 formation, Figure 1E, which is the reaction described in Equation (3). After elimination of H2O2 from Figure 1E the semiquinone is minimized. Hence, this polyphenol can interact with an additional superoxide, van der Waals posed π-π to the centroid ring (3.50 Å), Figure 1F. When this initial configuration is DFT geometrically optimized, Figure 1G, the result differs markedly from the σ attack by the superoxide in Figure 1C. In fact, the unpaired superoxide electron gets transferred into the ring and the formed molecule of O2 is displaced 5.989 Å, Figure 1G. This polyphenol anion reacts easily with an additional proton to reform the coumarin, Figure 1H. All these reactions proceed without any energy barrier. Therefore, the overall reaction involves 2 superoxide anions (Figure 1B,F) plus 2 protons (Figure 1D,H), and results in formation of H2O2 (Figure 1E) and O2 (Figure 1G), as well as coumarin reformation (Figure 1H), ready for a further cycle of scavenging. The overall sequence is represented in Equation (5), which is the same shown by the SOD enzymes.2 O2•- + 2H+ → O2 + H2O2 (5)
3. Galangin
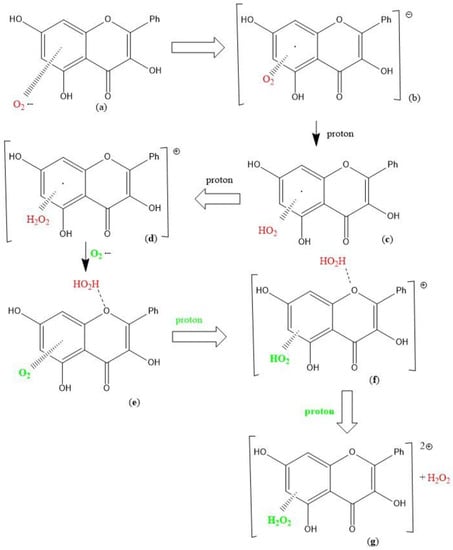
A recent study by us has shown the scavenging of superoxide by the flavonoid galangin [13]. As seen with the coumarin earlier described, a straightforward SOD action is observed when the first attack of superoxide occurs on galangin H3 and is shown in Scheme 1 below.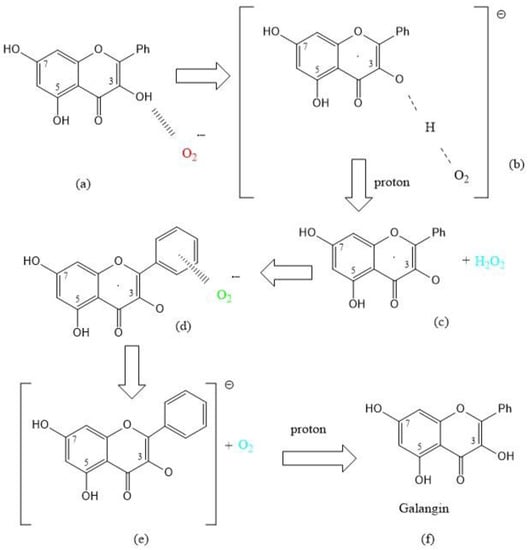
4. Butein
A study of butein and other related chalcone antioxidants was recently described by us [14]. RRDE cyclic voltammetry and X-ray diffraction data, followed by DFT and docking calculations show antioxidant and potential antimalarial properties. As shown by other antioxidants described in this work, butein (Figure 3A) is able to scavenge superoxide. The sequence of superoxide interaction starts with the butein H(hydroxyl) group in position 4 (ring B), Figure 3B. The whole sequence is shown in Figure 3A–F and is closely related to what has been above described for 4-methyl-7,8-di-hydroxy coumarin and galangin (Scheme 1).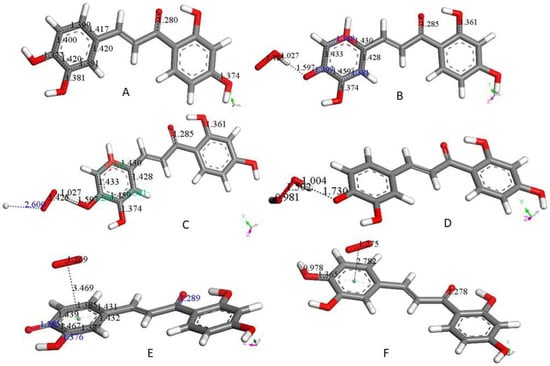
5. Additional Scavengers
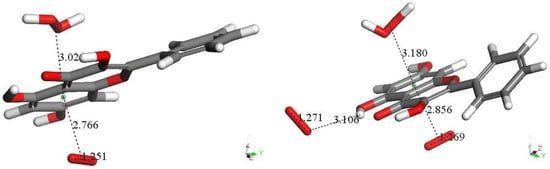
In this section, we study only the initial superoxide π-π interaction on other polyphenols. That is, this action precludes the most common superoxide σ attack on an H(hydroxyl). Figure 4 shows the DFT results after posing a superoxide (having O–O bond length of 1.373 Å) at 3.50 Å from a ring centroid. In Figure 4A, the energy-minimized 4-methyl-7,8-dimethoxy-coumarin structure [12] shows that the superoxide penetrates the ring environment, with centroid separation of 3.042 Å, while the superoxide O–O bond length becomes shorter, 1.323 Å, and so a coumarin-η-superoxide complex is formed. As seen in Figure 4B, a similar complex is reached by DFT minimization of the dimethoxy coumarin fraxidin [15], with even shorter separation between centroids, 2.927 Å. We were also interested in investigating potential superoxide interaction with the non-pyrone ring of fraxidin, Figure 4C. This interaction between both species is weak, as shown by separation between centroids of 3.348 Å, closer to the initial van der Waals separation. Hence, we studied the interaction between the pyrone ring of another coumarin, bergamottin [15]. The related bergamottin-η-superoxide complex shows a strong interaction with superoxide, as the distance between them is 2.955 Å, Figure 4D. Additional π-π complexes with superoxide are formed by the antioxidants galangin, Figure 4E, and 4-methyl,3,6,8-triacetate-coumarin [12], Figure 4F. These interactions (3.190 Å and 3.162 Å, respectively) are somewhat weaker than that of bergamottin and more similar to the superoxide-η-4-methyl-7,8-dimethoxy-coumarin complex. Butein here is an exception to compounds shown in Figure 4, where the π-π attack of superoxide is directed towards H(hydroxyl) in position 4, and so it is equivalent to the σ mechanism of scavenging shown in Figure 1B and Figure 3B. The related sequence is shown as a video in manuscript Supplementary Material, Video S1.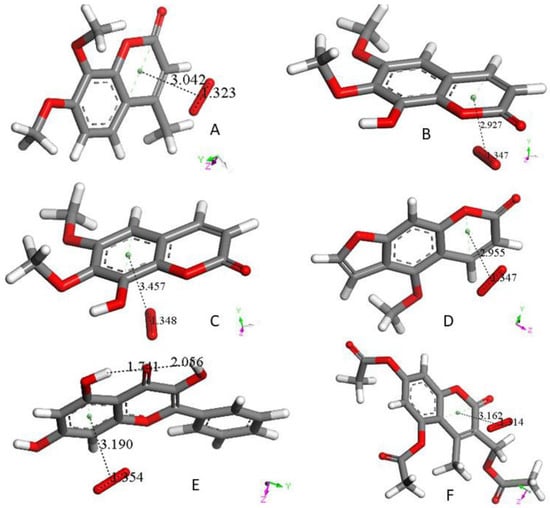
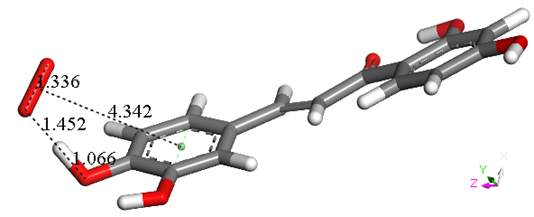
Figure 5. Geometry optimization of van der Waals π-π interaction between butein ring B and superoxide. Initial separation of 3.50 Å, evolves towards σ scavenging, in contrast with all figures in this section (Figure 4). The picture shows the evolution of the DFT process after 100 cycles of geometry optimization, clearly indicating that the result is the capture of H4 by superoxide, confirmed after additional cycles (not shown).
6. RRDE Cyclovoltammetry
Most of the studied compounds described in this work have also been studied using the Rotating Ring Disk Electrode method. This was developed in our lab [16] and resulted in a quantitative description for polyphenol scavenging of the superoxide radical. In an RRDE voltammetry experiment, the generation of the superoxide radicals occurs at the disk electrode, while the oxidation of the residual superoxide radicals (that have not been scavenged by the antioxidant) occurs at the ring electrode. Reaction 1: Reduction of molecular oxygen at disk electrodeTable 1 shows results according to this method: It is observed that catechol polyphenols are more effective than those having isolated single hydroxyl groups.
Table 1. Slopes of Collection Efficiency (indicators of scavenging superoxide) [16] for several polyphenols using the RRDE method.
|
BHT |
Chrysin |
Eriodictyol |
DHDM |
Butein |
Clovamide |
Quercetin ** |
Galangin |
|
-0.16x104 [17] |
-1.10x104 [16]
|
-2.20x104 [16]
|
−8.0x104 [14]
|
−11.2x104 [14]
|
−12.0x104 [18] |
−5.30x104 [16] −15.4x104 [17] −15.5x104 [18]
|
-19.0x104 [13] |
(**) Ref [16] has the earliest RRDE determination of quercetin, a polyphenol used as a standard for comparison with each RRDE scavenger analyzed by us. Quercetin in DMSO solutions tends to slightly decrease its yellowish color and its scavenging activity with time. Consequently, all successive RRDE studies were done using fresh solutions. Quercetin measured in this manner [17,18] shows the correct slope, steeper than in [16].
7. Conclusions
Polyphenols are valuable natural products present in our diet that may contribute to decrease aging effects and some diseases. However, because of their poor absorption in the gut and consequent low concentration in biological fluids, reservations about antioxidant polyphenol efficiency have been raised. In this study, we focus our attention on the potential reformation of polyphenols after scavenging superoxide and we find that polyphenol mimicking SOD enzyme activity is feasible for several polyphenolic compounds. Thus, after scavenging superoxide, 4-methyl-7,8-di-hydroxy-coumarin and galangin (Chart 1) exactly reform themselves; butein incorporates a molecule of O2 in the polyphenol system, e.g., a η-O2-butein complex is formed; galangin alternative mechanism (Chart 2) includes an additional H2O2 ligand and forms the η-(H2O2)-galangin-η-O2 complex.
We demonstrate using DFT methods that the π-π interaction between superoxide and one polyphenol ring is associated with oxidation of the superoxide radical, due to transfer of its unpaired electron to an aromatic ring of the polyphenol. This is also responsible for O2 release as part of SOD activity here proposed. However, it is very difficult to establish if this π-π reaction
O2•- + polyphenol-OH → O2 + polyphenol-OH•-
proceeds before or after the most common mechanism of polyphenol scavenging (abstraction of H(hydroxyl), σ scavenging)
O2•- + polyphenol-OH → HO2- + polyphenol-O•
In addition, the initial direct π-π interaction between superoxide and one polyphenol aromatic ring was also studied for a variety of polyphenols. In this specific initial π-π interaction, all polyphenols studied are shown through DFT methods to have minimum energy radical complexes of the type polyphenol-η-O2 except for butein. By circumventing the π-π interaction butein resulted in behaving as a typical σ scavenger of superoxide, e.g. sequestering a H(hydroxyl). It is suggested that these polyphenols are able to mimic SOD enzyme action in the disproportionation reaction of O2•- [equation (5)] and, thus refute the claim that low polyphenol concentration in biological fluids as a limiting factor in their effective scavenging of superoxide.
References
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res. 2018, 122, 877–902. [CrossRef] [PubMed]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [CrossRef]
- Bae, Y.S.; Oh, H.; Rhee, S.G.; Yoo, Y.D. Regulation of reactive oxygen species generation in cell signaling. Mol. Cells 2011, 32, 491–509. [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [CrossRef] [PubMed]
- Giacco, F.; Brownlee, M. Oxidative stress and diabetic complications. Circ. Res. 2010, 107, 1058–1070. [CrossRef]
- Maier, C.M.; Chan, P.H. Role of superoxide dismutases in oxidative damage and neurodegenerative disorders. Neuroscientist 2002, 8, 323–334. [CrossRef]
- Wu, J. Tackle the free radicals damage in COVID-19. Nitric Oxide 2020, 102, 39–41. [CrossRef]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 1995, 64, 97–112. [CrossRef] [PubMed]
- Andreyev, A.Y.; Kushnareva, Y.E.; Starkov, A.A. Mitochondrial metabolism of reactive oxygen species. Biochem. Mosc. 2005, 70, 200–214. [CrossRef]
- Jadresko, D.; Milicevi´c, A.; Jovanovic, I.N. Reactivity of flavonoids toward superoxide radical: An electrochemical approach. Electrochim. Acta 2022, 421, 140501. [CrossRef]
- Pedersen, J.Z.; Oliveira, C.; Incerpi, S.; Kumar, V.; Fiore, A.M.; De Vito, P.; Prasad, A.K.; Malhotra, S.V.; Parmar, V.S.; Saso, L. Antioxidant activity of 4-methylcoumarins. J. Pharm. Pharmacol. 2007, 59, 1721–1728. [CrossRef] [PubMed]
- Barzegar, A.; Davari, M.D.; Chaparzadeh, N.; Zarghami, N.; Pedersen, J.Z.; Incerpi, S.; Saso, L.; Moosavi-Movahedi, A.A. Theoretical and experimental studies on the structure-antioxidant activity relationship of synthetic 4-methylcoumarins. J. Iran. Chem. Soc. 2011, 8, 973–982. [CrossRef]
- Caruso, F.; Berinato, M.; Hernandez, M.; Belli, S.; Smart, C.; Rossi, M. Antioxidant properties of bee propolis and an important component, galangin, described by X-ray crystal structure, DFT-D and hydrodynamic voltammetry. PLoS ONE 2022, 17, e0267624. [CrossRef]
- Okoye, I.; Yu, S.; Caruso, F.; Rossi, M. X-ray Structure Determination, Antioxidant Voltammetry Studies of Butein and 2,4- Dihydroxy-3,4-dimethoxychalcone. Computational Studies of 4 Structurally Related 2,4-diOH Chalcones to Examine Their Antimalarial Activity by Binding to Falcipain-2. Molecules 2021, 26, 6511. [CrossRef] [PubMed]
- Rossi, M.; Aktar, S.; Davis, M.; Hefter Feuss, E.; Roman-Holba, S.; Wen, K.; Gahn, C.; Caruso, F. The Grapefruit Effect: Interaction between Cytochrome P450 and Coumarin Food Components, Bergamottin, Fraxidin and Osthole. X-ray Crystal Structure and DFT Studies. Molecules 2020, 25, 3158. [CrossRef] [PubMed]
- Belli, S.; Rossi, M.; Molasky, N.; Middleton, L.; Caldwell, C.; Bartow-McKenney, C.; Duong, M.; Chiu, J.; Gibbs, E.; Caldwell, A.; et al. Effective and Novel Application of Hydrodynamic Voltammetry to the Study of Superoxide Radical Scavenging by Natural Phenolic Antioxidants. Antioxidants 2019, 8, 14. [CrossRef]
- Caruso, F.; Rossi, M.; Kaur, S.; Garcia-Villar, E.; Molasky, N.; Belli, S.; Sitek, J.D.; Gionfra, F.; Pedersen, J.Z.; Incerpi, S. Antioxidant Properties of Embelin in Cell Culture. Electrochemistry and Theoretical Mechanism of Scavenging. Potential Scavenging of Superoxide Radical through the Cell Membrane. Antioxidants 2020, 9, 382. [CrossRef] [PubMed]
- Ye, N.; Belli, S.; Caruso, F.; Roy, G.; Rossi, M. Antioxidant studies by hydrodynamic voltammetry and DFT, quantitative analyses by HPLC-DAD of clovamide, a natural phenolic compound found in Theobroma Cacao L. beans. Food Chem. 2021, 341 Pt 2, 128260. [CrossRef] [PubMed]
