Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Boren Tian.
Osteosarcoma (OS) is a malignancy that is becoming increasingly common in adolescents. OS stem cells (OSCs) form a dynamic subset of OS cells that are responsible for malignant progression and chemoradiotherapy resistance. The unique properties of OSCs, including self-renewal, multilineage differentiation and metastatic potential, 149 depend closely on their tumor microenvironment.
- osteosarcoma
- cancer stem cell
- cell plastic
1. Introduction
Osteosarcoma (OS) is a malignancy that most commonly occurs in children and adolescents and is the second highest cause of cancer-related mortality in these groups [1,2,3][1][2][3]. There has been a rise in the annual incidence rate of OS to three cases per million individuals [4]. The majority of OS cases arises in the metaphyseal regions adjacent to the physis, including the distal femur, proximal tibia and the proximal humerus, with a strong capacity for proliferation [5]. Over the past 30 years, the treatment of OS has improved little, such that surgery accompanied with chemoradiotherapy remain as the main method of treatment [6]. Although novel clinical strategies such as gene editing, individualized treatment and novel molecular-targeted therapies, e.g., angiogenesis inhibitors, tyrosine kinase inhibitors and monoclonal antibodies, have all been deployed against OS, the outcomes for patients are poor, particularly those with more aggressive forms of the cancers [3,4,5,6,7][3][4][5][6][7]. Therefore, novel treatment strategies are in demand in clinical practice. In addition, the molecular mechanism underlying tumorigenesis and malignant metastasis needs to be studied in detail.
Based on the present research, OS is speculated to have two main origins, bone mesenchymal stem cells (BMSCs) and osteoblasts [8,9,10][8][9][10]. p53 as a classic cancer suppressor gene plays a key role in OS progression. The deficiency of p53 is an important reason leading to primary OS. In addition, retinoblastoma gene (Rb), cyclin dependent kinase inhibitor 2 (CDKN2), KRAS and c-Met also participate in the regulation of OS progression [8,9,10,11][8][9][10][11]. Within cancer tissues, there exist several dynamic subsets of cancer cells considered to be cancer stem cells (CSCs) or stem cell-like cancer cells [12,13][12][13]. CSCs have been frequently reported to exhibit stem cell properties and capabilities of long-term clonal proliferation, tumorigenicity, facilitating metastasis and promoting resistance to chemotherapy and radiotherapy [14,15][14][15]. Therefore, exploring the origins of cancer initiation and metastasis will likely facilitate the development of future therapies. In 1994, Lapidot et.al first reported that, in human acute myeloid leukemia, a rare population of CSCs exists [16]. Subsequently, an accumulating number of studies have also reported the existence of CSCs in other solid tumors, including prostate, glioblastoma, hepatoma, breast cancer and OS [17,18,19,20,21,22][17][18][19][20][21][22]. In fact, all types of malignant tumors consist of different subpopulations of tumor cells, leading to high degrees of heterogeneity.
The niche in which CSCs reside is the tumor microenvironment, where they co-exist with adjacent supporting cells, micro-vessels and the extracellular matrix [19,20][19][20]. In addition, the tumor microenvironment can contain soluble factors, such as chemokines and cytokines, whilst being under the influence of various mechanical factors, including matrix stiffness, solid stress and fluid stress [23,24][23][24]. In the OS microenvironment, OS stem cells (OSCs) are contained in a specialized niche that contains a unique bone microenvironment, which consists of various types of bone cells, such as osteoblasts or osteoclasts. OSCs are similar to other CSCs, in that they account for a proportion of cancer cells with tumorigenic and self-renewal capabilities. The existence of OSCs was first confirmed by Gibbs et al., who found that when primary human OS cells or the OS cell line MG63 were suspended in a serum-free medium with defined growth factors, 0.1% of the cells could form spheres with self-renewal capacity [17,18,19,20,21,22,23,24,25][17][18][19][20][21][22][23][24][25]. Subsequently, a series of studies have proven the existence of OSCs, in addition to revealing the phenotype and possible marker profile of OSCs. The recent studies on possible OSC markers and phenotypes (Table 1).
Table 1. Putative OSC markers and phenotypes.
| Marker | Cell Origin | Phenotype | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD133 | Saos-2, MG-63, U2-OS, MNNG/HOS, 143B, HOS, Human primary cells | High stem cells gene expression, sphere formation, side population, increased cell proliferation [26,27,28,29,30]. | High stem cells gene expression, sphere formation, side population, increased cell proliferation [26][27][28][29][30]. | ||||||||||||||||||||
| CD117/Stro-1 | K7M2, KHOS/NP, MNNG/HOS, 318–1, P932, BCOS | High stem cells gene expression, sphere formation, drug resistance, in vivo tumorigenicity and metastatic potential [17,18,19,20,21,22,23,24,25,26,27,28,29,30,31]. | High stem cells gene expression, sphere formation, drug resistance, in vivo tumorigenicity and metastatic potential [17][18][19][20][21][22][23][24][25][26][27][28][29][30][31]. | ||||||||||||||||||||
| CD271 | Human primary (FFPE), MNNG/HOS, U2-OS, Saos-2 | High stem cells gene expression, sphere formation, drug resistance, in vivo tumorigenicity [32]. | |||||||||||||||||||||
| Aldehyde dehydrogenase | MG-63, OS99–1 Hu09, Saos-2 | High stem cells gene expression, sphere formation, drug resistance, increased cell proliferation [33,34]. | High stem cells gene expression, sphere formation, drug resistance, increased cell proliferation [33][34]. | ||||||||||||||||||||
| Stem cells antigen-1 | 4 Murine osteosarcoma cell lines | Sphere formation, in vivo tumorigenicity [35,36] | Sphere formation, in vivo tumorigenicity [35][36] | ||||||||||||||||||||
| Fas apoptotic inhibitory molecule 2 | MNNG/HOS, U2-OS | Sphere formation, drug resistance, in vivo tumorigenicity [37]. | |||||||||||||||||||||
| Side population | OS2000, KIKU, NY, Huo9, HOS, U2OS, Saos-2, human primary |
High stem cells gene expression, Sphere formation, in vivo tumorigenicity, self-renewal, apoptosis resistant [38,39,40]. | High stem cells gene expression, Sphere formation, in vivo tumorigenicity, self-renewal, apoptosis resistant [38][39][40]. | ||||||||||||||||||||
| Sphere formation | MG-63, MNNG/HOS, human primary | High stem cells gene expression, drug resistance, in vivo tumorigenicity [17,18,19,20,21,22, | ][21 | 23, | ][22 | 24, | ][23 | 25, | ][24 | 32,33,41,42]. | High stem cells gene expression, drug resistance, in vivo tumorigenicity [17] | 26, | [18]][25 | 27, | [19] | 28, | [][ | 29, | 26][ | 30, | 2027] | 31, | [28][29][30][31][32][33][41][42]. |
2. Role of the Tumor Microenvironment in Regulating OSC Stemness
OSCs can interact with their microenvironment through complex and dynamic processes, including variation of oxygen, mechanical interactions, enzymatic modification of the extracellular matrix (ECM) structure and signaling cross-talk, all of which can influence the progression and the dissemination of OS cells (Figure 1)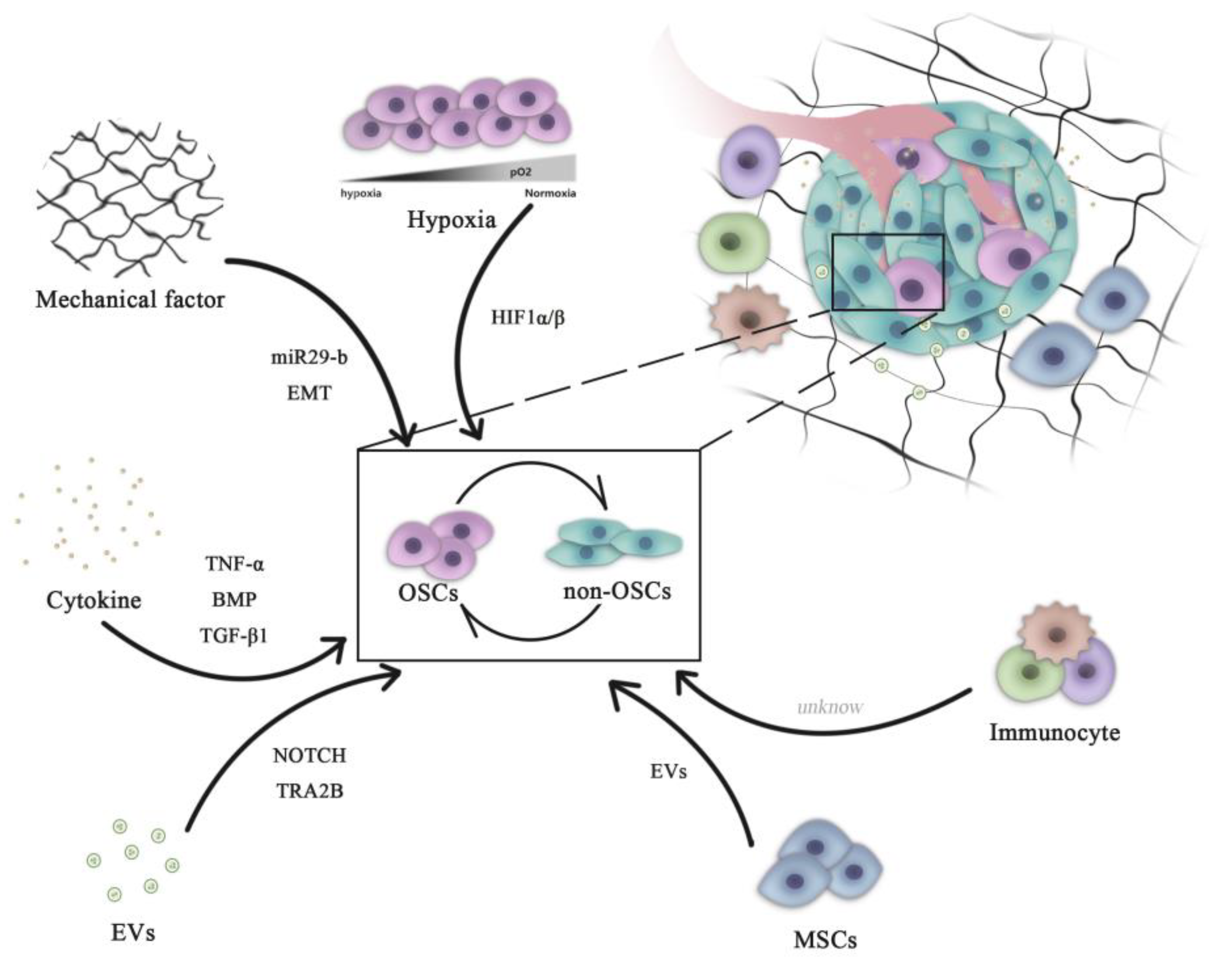
Figure 1. Role of microenvironment signaling in regulating OSC and non-OSC reversion. Complex pathways are necessary for the maintenance of the homeostasis of the OSC population. All microenvironment components, including cells (mesenchymal cells and immune cells) and non-cellular factors (hypoxia, cytokines and mechanical EVs), can influence the dynamic transition between OSCs and non-OSCs. OSCs, osteosarcoma stem cells; miR, microRNA; HIF, hypoxia-inducible factor; EVs, extracellular vesicles; EMT, epithelial-mesenchymal transition; BMP, bone morphogenetic protein; TNF, tumor necrosis factor; TGF, transforming growth factor; TRA2B, transformer 2β homolog.
2.1. Hypoxia
Common hallmarks of solid tumors include intra-tumoral hypoxia, necrosis, acidic environments and disturbed angiogenesis. Previous studies have shown that increased cancer cell stemness is associated with intra-tumoral hypoxia [54,55,56,57][43][44][45][46]. OurThe previous study demonstrated that a hypoxia microenvironment could induce non-OSCs dedifferentiation into OSCs by increasing the expression of TGF-β [58][47]. During the process of tumor formation, excessive proliferation of cancer cells consumes large quantities of oxygen in the microenvironment, resulting in the formation of a hypoxic zone in the central area of the tumor. In addition, aberrant secretion of angiogenetic factors, including vascular endothelial growth factor A and fibroblast growth factor 2 (FGF2), result in malformation and disorder in the neovascularization system [54,55][43][44]. This in turn causes the loss of oxygen supply, further aggravating hypoxia in the cancer tissue [59,60][48][49]. This hypoxic microenvironment induces the expression of hypoxia-inducible factor-1 (HIF-1), a vital member of the HIF family. By contrast, in the presence of oxygen, HIF-1 undergoes degradation by the von Hippel-Lindau protein, a tumor suppressor protein [61,62,63,64,65][50][51][52][53][54]. HIF-1 is a heterodimer that is ubiquitously expressed in human and mouse tissues. HIF-1 consists of two subunits, the hypoxia-inducible, oxygen-dependent subunit HIF-1α and the constitutively-expressed oxygen-independent subunit HIF-1β [66,67,68][55][56][57]. It is only when the oxygen concentration reaches <5% (such as when the volume of the tumor has grown to >300 mm2) that HIF-1α can exist stably. Activity of HIF-1 provides cancer cells with the ability to adapt to hypoxia and is closely associated with tumor metabolism, differentiation, angiogenesis, cell proliferation, metastasis and multidrug resistance. Of note, several studies have demonstrated that elevated expression of HIF-1α promoted the dedifferentiation of cancer cells into CSCs, whereas hypoxia is directly associated with poorer prognoses in patients with OS [69][58]. Numerous studies have demonstrated that hypoxia can promote the expression of the stem cell marker CD133 to maintain stemness and drug resistance in the Saos-2 OS cell line [70][59]. Lin et al. previously reported that hypoxia can increase the expression of embryonic stem cell markers, including Oct3/4 and Nanog, in the MNNG/HOS OS cell line [71][60]. Zhang et al. showed that a hypoxic microenvironment stabilized HIF-1α in OS cells, such that HIF-1 promoted the expression of microRNA (miR or miRNA)-210, which then induced and accelerated the dedifferentiation of OS cells into OSCs [58,59,60,61,62,63,64,65,66,67,68,69,70,71,72][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61]. These observations aforementioned suggest that HIF-1 and subsequent hypoxia signaling pathways can regulate the differentiation of CSCs and the dedifferentiation of non-stem cells in tumors [73,74][62][63]. In addition to HIF, hypoxia can also cause integrin-linked kinase dysfunction, triggering CSCs formation [75][64]. Hypoxia has been previously found to promote breast cancer stemness by HIF-dependent and AlkB homolog 5-mediated N6-methyladenosine (m6A)-demethylation of Nanog mRNA [76][65]. Shi et al. used the evolutionary theory to identify the hypoxic adaptation-associated gene YTH N6-methyladenosine RNA binding protein 1 (YTHDF1). As a member of the N6-methyladenosine (m6A)-modified RNA-binding protein family, YTHDF1 may interplay with other m6A modifiers and serve a pivotal role in the self-renewal and differentiation of stem cells [77][66]. Under hypoxia, AKT will accumulate in the mitochondria of tumor cells, whereby 3-phosphoinositide-dependent protein kinase 1 is phosphorylated at special sites. This pathway shifts the tumor metabolic program to glycolysis, which antagonizes apoptosis and autophagy and inhibits oxidative stress. This in turn maintains the survival and proliferation capabilities of tumor cells, as evidenced by the sphere-forming ability of cells in 3D cultures under severe hypoxia [78][67]. These aforementioned findings suggest that hypoxia may contribute to the creation of a microenvironment rich in tumor stem cells, where this unique hypoxic microenvironment may provide essential cellular interactions and environmental signals for the maintenance of CSCs [70,71,72,73,74,75,76,77,78,79][59][60][61][62][63][64][65][66][67][68]. By contrast, the hypoxia microenvironment can also regulate non-CSC dedifferentiation by regulating the activities of other pathways, including epithelial-mesenchymal transition (EMT), metabolic reprogramming, DNA hypermethylation and apoptotic resistance. Additionally, the hypoxic microenvironment can mediate the resistance of CSCs against drugs through drug transporters [80][69]. The majority of CSCs express the ATP-binding cassette (ABC) family of membrane transporters at high levels, including multidrug resistance gene 1, breast cancer resistance protein and multidrug resistance-associated protein. These proteins can transport metabolites, drugs and other substances, allowing CSCs to become highly resistant to chemotherapy. The relationship between hypoxia and ABC proteins was previously documented to have a strong association with mediating tumor drug resistance [81][70]. In conclusion, the hypoxic microenvironment with the activation of hypoxic signaling can serve key roles in the dedifferentiation of OS cells into OSCs. Therefore, it is important to study the molecular mechanism underlying OS dedifferentiation, which is expected to hold important clinical significance for improving the efficacy of therapeutic strategies. However, the molecular mechanism of how exactly the hypoxic microenvironment can regulate OSC physiology biology requires additional experimental evidence for validation. In particular, HIF-1 is a key molecule of the hypoxia signaling pathway, the downstream molecules of which are expected to become important markers and potential molecular targets of OSCs.2.2. Biomechanical Force
Under physiological conditions, most if not all organisms experience complex biomechanical forces, including shear stress, matrix stiffness, tension and compression pressure [82,83,84,85,86][71][72][73][74][75]. Biomechanical forces experienced by solid tumors have different profiles compare with those in the surrounding or healthy tissue [87][76]. Throughout the process of cancer development, excessive cell proliferation will lead to the abnormal development of the biomechanical microenvironment, including solid stress, increased matrix stiffness (decrease in OS due to osteolysis) and abnormal interstitial fluid pressure [88,89,90][77][78][79]. These complex mechanical systems are essential for the maintenance of the homeostasis of the CSC population. Indeed, previous studies have demonstrated that non-CSCs can be transformed into CSCs by receiving mechanical signals from the surrounding microenvironment, such as increased matrix stiffness [91,92,93,94][80][81][82][83] and/or fluid shear stress [95,96,97,98][84][85][86][87]. In OS, soft substrate (7 kPa) has been reported to preserve OS stemness, mainly through miR-29b/Spin 1-dependent signaling. Manipulation of cancer niche stiffness and miR-29b expression may therefore be potentially novel drug targets in OS [99][88]. Previous studies have shown that EMT can promote the progression and invasion of tumors [100][89]. EMT have been observed to serve as a direct link between non-CSCs and the gain of CSC properties [101][90]. Matrix stiffness in the tumor microenvironment can actively regulate EMT and migration of OS cells through cytoskeletal remodeling and the translocation of myocardin related transcription factor A, which may contribute to cancer progression [102][91]. Although the aforementioned studies revealed that mechanical factors are at least partially associated with the dynamic conversion between non-CSCs and CSCs, further research into the association between mechanical signaling and OSC stemness is warranted. Mechanical receptors on the cell surface, such as integrins, CD44 and ion channels, can sense the changes in ECM and activate key downstream molecules, including focal adhesion kinase, integrin-linked kinase, RhoA and yes-associated protein. Several of the signals induce non-CSC reprogramming and transform them into CSCs by increasing the expression of sex determining region Y-box 2 (Sox2), octamer-binding transcription factor (Oct)-4 and Nanog [91,92,93,94,95,96,97,98,99,100,101,102,103,104][80][81][82][83][84][85][86][87][88][89][90][91][92][93]. CSCs and normal stem cells frequently share similar surface markers and signaling pathways, which would restrict the design of treatment regimens [105][94]. The abnormal mechanical system in OS microenvironments, which rarely occur in the harmonious microenvironments of normal stem cells, may provide novel insights for designing CSC-targeted treatment methods. As such, discovering the relationship between biomechanical factors and CSCs will greatly enable the generation of novel research strategies to investigate the occurrence, development, and recurrence of cancers.2.3. Growth Factors
Growth factors are pivotal in maintaining the physiological behavior of healthy individuals. Cells in the tumor microenvironment can secrete growth factors to regulate processes of tumor development [106,107][95][96]. When OS arise in the bone, OS cells secrete factors that direct osteoclast-mediated bone destruction. In addition, matrix-derived growth factors, especially transforming growth factor β1 (TGF-β1), are released from bone matrix. In addition, OS cells can release TGF-β1 directly, where increased TGF-β1 expression is associated with high-grade metastases of OS [108][97]. TGF-β1 is a multi-function cytokine that serves as a mediator in the tumor to facilitate further tumor expansion, metastasis and cytokine production [109][98]. Wang et.al previously reported that TGF-β1 can switch the OSC chemoresistance through the miR-499a/SHKBP1 axis [110][99]. In another study, TGF-β1 signaling and a hypoxic environment were found to induce the transformation of non-OSCs into OSCs dynamically, which promoted the acquisition of chemoresistance, tumorigenicity, neovasculo-genicity and metastatic potential. Furthermore, blocking the TGF-β1 signaling pathway was reported to inhibit this switch from non-OSCs to OSCs, inhibit OSC self-renewal and suppress hypoxia-mediated dedifferentiation [58][47]. In the bone microenvironment, TGF-β1 signaling is responsible for OSC generation and critical to chemoresistance in vivo. In addition to OS, TGF-β1 can also regulate the dynamic switching between stem cells and non-stem cells to influence the progression of tumors from different tissue origins [111,112][100][101]. In conclusion, TGF-β1 serves a key role in regulating the dynamic plasticity of OSC, which can lead to non-stem cells adopting OSC characteristics to promote tumorigenesis and chemoresistance, highlighting TGF-β1 as a potential therapeutic target. Bone morphogenetic proteins (BMPs) are members of the TGF-β superfamily and serve important roles in the activity of various tissues. In OS, BMP-2 suppresses tumor growth by reducing the expression of oncogenes whilst promoting the differentiation of OSCs [113][102]. Histological examination and gene expression analysis of OS tissues revealed that fibrotic remodeling of the tumor microenvironment favors tumorigenesis. Zhang et al. previously demonstrated that fibrotic reprogramming in the lung induced by OSCs is critical for OS pulmonary metastasis, with FGF-FGF receptor 2 (FGFR2) signaling being responsible for this important process [114][103]. In OS, the tumor necrosis factor-α/miR-155 axis has been found to induce OSC transformation between non-OSCs and OSCs through the extracellular signal-regulated protein kinase signaling pathway [115][104]. Melatonin, one of the hormones secreted by the pineal gland of the brain, has been shown to significantly inhibit sphere formation by OSCs through the key transcription factor Sox-9 [116][105]. Although all of the aforementioned growth factors have shown the potential to target OSCs, the underlying mechanism require further exploration.References
- Turcotte, L.M.; Neglia, J.P.; Reulen, R.C.; Ronckers, C.M.; van Leeuwen, F.E.; Morton, L.M.; Hodgson, D.C.; Yasui, Y.; Oeffinger, K.C.; Henderson, T.O. Risk, Risk Factors, and Surveillance of Subsequent Malignant Neoplasms in Survivors of Childhood Cancer: A Review. J. Clin. Oncol. 2018, 36, 2145–2152.
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Prim. 2018, 4, 5.
- Smith, R.A.; Andrews, K.S.; Brooks, D.; Fedewa, S.A.; Manassaram-Baptiste, D.; Saslow, D.; Wender, R.C. Cancer screening in the United States, 2019: A review of current American Cancer Society guidelines and current issues in cancer screening. CA A Cancer J. Clin. 2019, 69, 184–210.
- Gianferante, D.M.; Mirabello, L.; Savage, S.A. Germline and somatic genetics of osteosarcoma—Connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 2017, 13, 480–491.
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W.; et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002, 20, 776–790.
- Crompton, J.G.; Ogura, K.; Bernthal, N.M.; Kawai, A.; Eilber, F.C. Local Control of Soft Tissue and Bone Sarcomas. J. Clin. Oncol. 2018, 36, 111–117.
- Buja, A.; Lago, L.; Lago, S.; Vinelli, A.; Zanardo, C.; Baldo, V. Marital status and stage of cancer at diagnosis: A systematic review. Eur. J. Cancer Care 2018, 27, e12755.
- Rubio, R.; Abarrategi, A.; Garcia-Castro, J.; Martinez-Cruzado, L.; Suarez, C.; Tornin, J.; Santos, L.; Astudillo, A.; Colmenero, I.; Mulero, F.; et al. Bone environment is essential for osteosarcoma development from transformed mesenchymal stem cells. Stem Cells 2014, 32, 1136–1148.
- Mohseny, A.B.; Szuhai, K.; Romeo, S.; Buddingh, E.P.; Briaire-de Bruijn, I.; de Jong, D.; van Pel, M.; Cleton-Jansen, A.M.; Hogendoorn, P.C. Osteosarcoma originates from mesenchymal stem cells in consequence of aneuploidization and genomic loss of Cdkn2. J. Pathol. 2009, 219, 294–305.
- Basu-Roy, U.; Basilico, C.; Mansukhani, A. Perspectives on cancer stem cells in osteosarcoma. Cancer Lett. 2013, 338, 158–167.
- Zhang, Y.; Mai, Q.; Zhang, X.; Xie, C.; Zhang, Y. Microenvironment Signals and Mechanisms in the Regulation of Osteosarcoma. Osteosarcoma Biol. Behav. Mech. 2017.
- Teng, Y.D.; Wang, L.; Kabatas, S.; Ulrich, H.; Zafonte, R.D. Cancer Stem Cells or Tumor Survival Cells? Stem Cells Dev. 2018, 27, 1466–1478.
- Medema, J.P. Cancer stem cells: The challenges ahead. Nat. Cell Biol. 2013, 15, 338–344.
- Arima, Y.; Nobusue, H.; Saya, H. Targeting of cancer stem cells by differentiation therapy. Cancer Sci. 2020, 111, 2689–2695.
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134.
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648.
- Gibbs, C.P.; Kukekov, V.G.; Reith, J.D.; Tchigrinova, O.; Suslov, O.N.; Scott, E.W.; Ghivizzani, S.C.; Ignatova, T.N.; Steindler, D.A. Stem-like cells in bone sarcomas: Implications for tumorigenesis. Neoplasia 2005, 7, 967–976.
- Klarmann, G.J.; Hurt, E.M.; Mathews, L.A.; Zhang, X.; Duhagon, M.A.; Mistree, T.; Thomas, S.B.; Farrar, W.L. Invasive prostate cancer cells are tumor initiating cells that have a stem cell-like genomic signature. Clin. Exp. Metastasis 2009, 26, 433–446.
- Yuan, X.; Curtin, J.; Xiong, Y.; Liu, G.; Waschsmann-Hogiu, S.; Farkas, D.L.; Black, K.L.; Yu, J.S. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 2004, 23, 9392–9400.
- Cao, L.; Zhou, Y.; Zhai, B.; Liao, J.; Xu, W.; Zhang, R.; Li, J.; Zhang, Y.; Chen, L.; Qian, H.; et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011, 11, 71.
- Ma, S.; Chan, K.W.; Hu, L.; Lee, T.K.; Wo, J.Y.; Ng, I.O.; Zheng, B.J.; Guan, X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556.
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005, 65, 5506–5511.
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238.
- Ju, F.; Atyah, M.M.; Horstmann, N.; Gul, S.; Vago, R.; Bruns, C.J.; Zhao, Y.; Dong, Q.Z.; Ren, N. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res. Ther. 2022, 13, 233.
- Gibbs, C.P., Jr.; Levings, P.P.; Ghivizzani, S.C. Evidence for the osteosarcoma stem cell. Curr. Orthop. Pract. 2011, 22, 322–326.
- Tirino, V.; Desiderio, V.; d’Aquino, R.; De Francesco, F.; Pirozzi, G.; Graziano, A.; Galderisi, U.; Cavaliere, C.; De Rosa, A.; Papaccio, G.; et al. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS ONE 2008, 3, e3469.
- Li, J.; Zhong, X.Y.; Li, Z.Y.; Cai, J.F.; Zou, L.; Li, J.M.; Yang, T.; Liu, W. CD133 expression in osteosarcoma and derivation of CD133(+) cells. Mol. Med. Rep. 2013, 7, 577–584.
- He, A.; Qi, W.; Huang, Y.; Feng, T.; Chen, J.; Sun, Y.; Shen, Z.; Yao, Y. CD133 expression predicts lung metastasis and poor prognosis in osteosarcoma patients: A clinical and experimental study. Exp. Ther. Med. 2012, 4, 435–441.
- Tirino, V.; Desiderio, V.; Paino, F.; De Rosa, A.; Papaccio, F.; Fazioli, F.; Pirozzi, G.; Papaccio, G. Human primary bone sarcomas contain CD133+ cancer stem cells displaying high tumorigenicity in vivo. FASEB J. 2011, 25, 2022–2030.
- Fujiwara, T.; Katsuda, T.; Hagiwara, K.; Kosaka, N.; Yoshioka, Y.; Takahashi, R.U.; Takeshita, F.; Kubota, D.; Kondo, T.; Ichikawa, H.; et al. Clinical relevance and therapeutic significance of microRNA-133a expression profiles and functions in malignant osteosarcoma-initiating cells. Stem Cells 2014, 32, 959–973.
- Adhikari, A.S.; Agarwal, N.; Wood, B.M.; Porretta, C.; Ruiz, B.; Pochampally, R.R.; Iwakuma, T. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010, 70, 4602–4612.
- Tian, J.; Li, X.; Si, M.; Liu, T.; Li, J. CD271+ osteosarcoma cells display stem-like properties. PLoS ONE 2014, 9, e98549.
- Honoki, K.; Fujii, H.; Kubo, A.; Kido, A.; Mori, T.; Tanaka, Y.; Tsujiuchi, T. Possible involvement of stem-like populations with elevated ALDH1 in sarcomas for chemotherapeutic drug resistance. Oncol. Rep. 2010, 24, 501–505.
- Wang, L.; Park, P.; Zhang, H.; La Marca, F.; Lin, C.Y. Prospective identification of tumorigenic osteosarcoma cancer stem cells in OS99-1 cells based on high aldehyde dehydrogenase activity. Int. J. Cancer 2011, 128, 294–303.
- Berman, S.D.; Calo, E.; Landman, A.S.; Danielian, P.S.; Miller, E.S.; West, J.C.; Fonhoue, B.D.; Caron, A.; Bronson, R.; Bouxsein, M.L.; et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc. Natl. Acad. Sci. USA 2008, 105, 11851–11856.
- Walkley, C.R.; Qudsi, R.; Sankaran, V.G.; Perry, J.A.; Gostissa, M.; Roth, S.I.; Rodda, S.J.; Snay, E.; Dunning, P.; Fahey, F.H.; et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008, 22, 1662–1676.
- Pan, Y.; Zhang, Y.; Tang, W.; Zhang, Y. Interstitial serum albumin empowers osteosarcoma cells with FAIM2 transcription to obtain viability via dedifferentiation. In Vitro Cell Dev. Biol. Anim. 2020, 56, 129–144.
- Murase, M.; Kano, M.; Tsukahara, T.; Takahashi, A.; Torigoe, T.; Kawaguchi, S.; Kimura, S.; Wada, T.; Uchihashi, Y.; Kondo, T.; et al. Side population cells have the characteristics of cancer stem-like cells/cancer-initiating cells in bone sarcomas. Br. J. Cancer 2009, 101, 1425–1432.
- Yang, M.; Yan, M.; Zhang, R.; Li, J.; Luo, Z. Side population cells isolated from human osteosarcoma are enriched with tumor-initiating cells. Cancer Sci. 2011, 102, 1774–1781.
- Wang, Y.; Teng, J.S. Increased multi-drug resistance and reduced apoptosis in osteosarcoma side population cells are crucial factors for tumor recurrence. Exp. Ther. Med. 2016, 12, 81–86.
- Rainusso, N.; Man, T.K.; Lau, C.C.; Hicks, J.; Shen, J.J.; Yu, A.; Wang, L.L.; Rosen, J.M. Identification and gene expression profiling of tumor-initiating cells isolated from human osteosarcoma cell lines in an orthotopic mouse model. Cancer Biol. 2011, 12, 278–287.
- Martins-Neves, S.R.; Corver, W.E.; Paiva-Oliveira, D.I.; van den Akker, B.E.; Briaire-de-Bruijn, I.H.; Bovee, J.V.; Gomes, C.M.; Cleton-Jansen, A.M. Osteosarcoma Stem Cells Have Active Wnt/beta-catenin and Overexpress SOX2 and KLF4. J. Cell. Physiol. 2016, 231, 876–886.
- Prasad, P.; Mittal, S.A.; Chongtham, J.; Mohanty, S.; Srivastava, T. Hypoxia-Mediated Epigenetic Regulation of Stemness in Brain Tumor Cells. Stem Cells 2017, 35, 1468–1478.
- Gilchrist, K.W.; Gray, R.; Fowble, B.; Tormey, D.C.; Taylor, S.G.t. Tumor necrosis is a prognostic predictor for early recurrence and death in lymph node-positive breast cancer: A 10-year follow-up study of 728 Eastern Cooperative Oncology Group patients. J. Clin. Oncol. 1993, 11, 1929–1935.
- Beck, B.; Driessens, G.; Goossens, S.; Youssef, K.K.; Kuchnio, A.; Caauwe, A.; Sotiropoulou, P.A.; Loges, S.; Lapouge, G.; Candi, A.; et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature 2011, 478, 399–403.
- Zhang, B.; Li, Y.L.; Zhao, J.L.; Zhen, O.; Yu, C.; Yang, B.H.; Yu, X.R. Hypoxia-inducible factor-1 promotes cancer progression through activating AKT/Cyclin D1 signaling pathway in osteosarcoma. Biomed. Pharm. 2018, 105, 1–9.
- Zhang, H.; Wu, H.; Zheng, J.; Yu, P.; Xu, L.; Jiang, P.; Gao, J.; Wang, H.; Zhang, Y. Transforming growth factor beta1 signal is crucial for dedifferentiation of cancer cells to cancer stem cells in osteosarcoma. Stem Cells 2013, 31, 433–446.
- Maes, C.; Carmeliet, G.; Schipani, E. Hypoxia-driven pathways in bone development, regeneration and disease. Nat. Rev. Rheumatol. 2012, 8, 358–366.
- Godet, I.; Shin, Y.J.; Ju, J.A.; Ye, I.C.; Wang, G.; Gilkes, D.M. Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis. Nat. Commun. 2019, 10, 4862.
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275.
- Vanharanta, S.; Shu, W.; Brenet, F.; Hakimi, A.A.; Heguy, A.; Viale, A.; Reuter, V.E.; Hsieh, J.J.; Scandura, J.M.; Massagué, J. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat. Med. 2013, 19, 50–56.
- Mekhail, K.; Gunaratnam, L.; Bonicalzi, M.E.; Lee, S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004, 6, 642–647.
- Giaccia, A.J.; Simon, M.C.; Johnson, R. The biology of hypoxia: The role of oxygen sensing in development, normal function, and disease. Genes Dev. 2004, 18, 2183–2194.
- Rankin, E.B.; Giaccia, A.J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008, 15, 678–685.
- Lee, J.W.; Bae, S.H.; Jeong, J.W.; Kim, S.H.; Kim, K.W. Hypoxia-inducible factor (HIF-1)alpha: Its protein stability and biological functions. Exp. Mol. Med. 2004, 36, 1–12.
- Ke, Q.; Costa, M. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480.
- Koh, M.Y.; Powis, G. Passing the baton: The HIF switch. Trends Biochem. Sci. 2012, 37, 364–372.
- Ouyang, Y.; Li, H.; Bu, J.; Li, X.; Chen, Z.; Xiao, T. Hypoxia-inducible factor-1 expression predicts osteosarcoma patients’ survival: A meta-analysis. Int. J. Biol. Mrk. 2016, 31, e229–e234.
- Koka, P.; Mundre, R.S.; Rangarajan, R.; Chandramohan, Y.; Subramanian, R.K.; Dhanasekaran, A. Uncoupling Warburg effect and stemness in CD133(+ve) cancer stem cells from Saos-2 (osteosarcoma) cell line under hypoxia. Mol. Biol. Rep. 2018, 45, 1653–1662.
- Lin, J.; Wang, X.; Wang, X.; Wang, S.; Shen, R.; Yang, Y.; Xu, J.; Lin, J. Hypoxia increases the expression of stem cell markers in human osteosarcoma cells. Oncol. Lett. 2021, 21, 217.
- Zhang, H.; Mai, Q.; Chen, J. MicroRNA-210 is increased and it is required for dedifferentiation of osteosarcoma cell line. Cell Biol. Int. 2017, 41, 267–275.
- Méndez, O.; Zavadil, J.; Esencay, M.; Lukyanov, Y.; Santovasi, D.; Wang, S.C.; Newcomb, E.W.; Zagzag, D. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol. Cancer 2010, 9, 133.
- Couvelard, A.; O’Toole, D.; Turley, H.; Leek, R.; Sauvanet, A.; Degott, C.; Ruszniewski, P.; Belghiti, J.; Harris, A.L.; Gatter, K.; et al. Microvascular density and hypoxia-inducible factor pathway in pancreatic endocrine tumours: Negative correlation of microvascular density and VEGF expression with tumour progression. Br. J. Cancer 2005, 92, 94–101.
- Pang, M.F.; Siedlik, M.J.; Han, S.; Stallings-Mann, M.; Radisky, D.C.; Nelson, C.M. Tissue Stiffness and Hypoxia Modulate the Integrin-Linked Kinase ILK to Control Breast Cancer Stem-like Cells. Cancer Res. 2016, 76, 5277–5287.
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056.
- Shi, Y.; Fan, S.; Wu, M.; Zuo, Z.; Li, X.; Jiang, L.; Shen, Q.; Xu, P.; Zeng, L.; Zhou, Y.; et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019, 10, 4892.
- Chae, Y.C.; Vaira, V.; Caino, M.C.; Tang, H.Y.; Seo, J.H.; Kossenkov, A.V.; Ottobrini, L.; Martelli, C.; Lucignani, G.; Bertolini, I.; et al. Mitochondrial Akt Regulation of Hypoxic Tumor Reprogramming. Cancer Cell 2016, 30, 257–272.
- Gorgun, C.; Ozturk, S.; Gokalp, S.; Vatansever, S.; Gurhan, S.I.; Urkmez, A.S. Synergistic role of three dimensional niche and hypoxia on conservation of cancer stem cell phenotype. Int. J. Biol. Macromol. 2016, 90, 20–26.
- Najafi, M.; Farhood, B.; Mortezaee, K.; Kharazinejad, E.; Majidpoor, J.; Ahadi, R. Hypoxia in solid tumors: A key promoter of cancer stem cell (CSC) resistance. J. Cancer Res. Clin. Oncol. 2020, 146, 19–31.
- Sun, X.; Lv, X.; Yan, Y.; Zhao, Y.; Ma, R.; He, M.; Wei, M. Hypoxia-mediated cancer stem cell resistance and targeted therapy. Biomed. Pharm. 2020, 130, 110623.
- Nathan, S.S.; DiResta, G.R.; Casas-Ganem, J.E.; Hoang, B.H.; Sowers, R.; Yang, R.; Huvos, A.G.; Gorlick, R.; Healey, J.H. Elevated physiologic tumor pressure promotes proliferation and chemosensitivity in human osteosarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 2389–2397.
- Pisanu, M.E.; Noto, A.; De Vitis, C.; Masiello, M.G.; Coluccia, P.; Proietti, S.; Giovagnoli, M.R.; Ricci, A.; Giarnieri, E.; Cucina, A.; et al. Lung cancer stem cell lose their stemness default state after exposure to microgravity. BioMed Res. Int. 2014, 2014, 470253.
- Lu, D.; Luo, C.; Zhang, C.; Li, Z.; Long, M. Differential regulation of morphology and stemness of mouse embryonic stem cells by substrate stiffness and topography. Biomaterials 2014, 35, 3945–3955.
- Hao, J.; Zhang, Y.; Ye, R.; Zheng, Y.; Zhao, Z.; Li, J. Mechanotransduction in cancer stem cells. Cell Biol. Int. 2013, 37, 888–891.
- Tian, B.; Lin, W.; Zhang, Y. Effects of biomechanical forces on the biological behavior of cancer stem cells. J. Cancer 2021, 12, 5895–5902.
- Northey, J.J.; Przybyla, L.; Weaver, V.M. Tissue Force Programs Cell Fate and Tumor Aggression. Cancer Discov. 2017, 7, 1224–1237.
- Shieh, A.C. Biomechanical forces shape the tumor microenvironment. Ann. Biomed. Eng. 2011, 39, 1379–1389.
- Mierke, C.T. The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells. Rep. Prog. Phys. Phys. Soc. 2019, 82, 064602.
- Plotkin, L.I.; Bellido, T. Osteocytic signalling pathways as therapeutic targets for bone fragility. Nat. Rev. Endocrinol. 2016, 12, 593–605.
- You, Y.; Zheng, Q.; Dong, Y.; Xie, X.; Wang, Y.; Wu, S.; Zhang, L.; Wang, Y.; Xue, T.; Wang, Z.; et al. Matrix stiffness-mediated effects on stemness characteristics occurring in HCC cells. Oncotarget 2016, 7, 32221–32231.
- Tan, F.; Huang, Y.; Pei, Q.; Liu, H.; Pei, H.; Zhu, H. Matrix stiffness mediates stemness characteristics via activating the Yes-associated protein in colorectal cancer cells. J. Cell. Biochem. 2018, 120, 2213–2225.
- Tian, B.; Luo, Q.; Ju, Y.; Song, G. A Soft Matrix Enhances the Cancer Stem Cell Phenotype of HCC Cells. Int. J. Mol. Sci. 2019, 20, 2831.
- Tan, Y.; Tajik, A.; Chen, J.; Jia, Q.; Chowdhury, F.; Wang, L.; Chen, J.; Zhang, S.; Hong, Y.; Yi, H.; et al. Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat. Commun. 2014, 5, 4619.
- Ip, C.K.; Li, S.S.; Tang, M.Y.; Sy, S.K.; Ren, Y.; Shum, H.C.; Wong, A.S. Stemness and chemoresistance in epithelial ovarian carcinoma cells under shear stress. Sci. Rep. 2016, 6, 26788.
- Triantafillu, U.L.; Park, S.; Klaassen, N.L.; Raddatz, A.D.; Kim, Y. Fluid shear stress induces cancer stem cell-like phenotype in MCF7 breast cancer cell line without inducing epithelial to mesenchymal transition. Int. J. Oncol. 2017, 50, 993–1001.
- Sun, J.; Luo, Q.; Liu, L.; Song, G. Low-level shear stress induces differentiation of liver cancer stem cells via the Wnt/beta-catenin signalling pathway. Exp. Cell Res. 2018, 397, 90–96.
- Sun, J.; Luo, Q.; Liu, L.; Song, G. Low-level shear stress promotes migration of liver cancer stem cells via the FAK-ERK1/2 signalling pathway. Cancer Lett. 2018, 427, 1–8.
- Li, S.; Bai, H.; Chen, X.; Gong, S.; Xiao, J.; Li, D.; Li, L.; Jiang, Y.; Li, T.; Qin, X.; et al. Soft Substrate Promotes Osteosarcoma Cell Self-Renewal, Differentiation, and Drug Resistance Through miR-29b and Its Target Protein Spin 1. ACS Biomater. Sci. Eng. 2020, 6, 5588–5598.
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711.
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715.
- Dai, J.; Qin, L.; Chen, Y.; Wang, H.; Lin, G.; Li, X.; Liao, H.; Fang, H. Matrix stiffness regulates epithelial-mesenchymal transition via cytoskeletal remodeling and MRTF-A translocation in osteosarcoma cells. J. Mech. Behav. Biomed. Mater. 2019, 90, 226–238.
- Gupta, R.K.; Johansson, S. beta1 integrins restrict the growth of foci and spheroids. Histochem. Cell Biol. 2012, 138, 881–894.
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205.
- Bu, Y.; Cao, D. The origin of cancer stem cells. Front. Biosci. 2012, 4, 819–830.
- Lowery, F.J.; Yu, D. Growth factor signaling in metastasis: Current understanding and future opportunities. Cancer Metastasis Rev. 2012, 31, 479–491.
- Lopez de Andres, J.; Grinan-Lison, C.; Jimenez, G.; Marchal, J.A. Cancer stem cell secretome in the tumor microenvironment: A key point for an effective personalized cancer treatment. J. Hematol. Oncol. 2020, 13, 136.
- Lamora, A.; Talbot, J.; Bougras, G.; Amiaud, J.; Leduc, M.; Chesneau, J.; Taurelle, J.; Stresing, V.; Le Deley, M.C.; Heymann, M.F.; et al. Overexpression of smad7 blocks primary tumor growth and lung metastasis development in osteosarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 5097–5112.
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-beta signal transduction for fibrosis and cancer therapy. Mol. Cancer 2022, 21, 104.
- Wang, T.; Wang, D.; Zhang, L.; Yang, P.; Wang, J.; Liu, Q.; Yan, F.; Lin, F. The TGFbeta-miR-499a-SHKBP1 pathway induces resistance to EGFR inhibitors in osteosarcoma cancer stem cell-like cells. J. Exp. Clin. Cancer Res. CR 2019, 38, 226.
- Matsumoto, T.; Yokoi, A.; Hashimura, M.; Oguri, Y.; Akiya, M.; Saegusa, M. TGF-β-mediated LEFTY/Akt/GSK-3β/Snail axis modulates epithelial-mesenchymal transition and cancer stem cell properties in ovarian clear cell carcinomas. Mol. Carcinog. 2018, 57, 957–967.
- Zhang, B.; Ye, H.; Ren, X.; Zheng, S.; Zhou, Q.; Chen, C.; Lin, Q.; Li, G.; Wei, L.; Fu, Z.; et al. Macrophage-expressed CD51 promotes cancer stem cell properties via the TGF-β1/smad2/3 axis in pancreatic cancer. Cancer Lett. 2019, 459, 204–215.
- Wang, L.; Park, P.; Zhang, H.; La Marca, F.; Claeson, A.; Valdivia, J.; Lin, C.-Y. BMP-2 inhibits the tumorigenicity of cancer stem cells in human osteosarcoma OS99-1 cell line. Cancer Biol. Ther. 2014, 11, 457–463.
- Zhang, W.; Zhao, J.M.; Lin, J.; Hu, C.Z.; Zhang, W.B.; Yang, W.L.; Zhang, J.; Zhang, J.W.; Zhu, J. Adaptive Fibrogenic Reprogramming of Osteosarcoma Stem Cells Promotes Metastatic Growth. Cell Rep. 2018, 24, 1266–1277.
- Yao, J.; Lin, J.; He, L.; Huang, J.; Liu, Q. TNF-alpha/miR-155 axis induces the transformation of osteosarcoma cancer stem cells independent of TP53INP1. Gene 2020, 726, 144224.
- Qu, H.; Xue, Y.; Lian, W.; Wang, C.; He, J.; Fu, Q.; Zhong, L.; Lin, N.; Lai, L.; Ye, Z.; et al. Melatonin inhibits osteosarcoma stem cells by suppressing SOX9-mediated signaling. Life Sci. 2018, 207, 253–264.
More
