Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Mahmood Barani and Version 2 by Rita Xu.
Klebsiella pneumoniae
is an important human pathogen that causes diseases such as urinary tract infections, pneumonia, bloodstream infections, bacteremia, and sepsis. The rise of multidrug-resistant strains has severely limited the available treatments for
K. pneumoniae
infections. On the other hand,
K. pneumoniae
activity (and related infections) urgently requires improved management strategies. A growing number of medical applications are using nanotechnology, which uses materials with atomic or molecular dimensions, to diagnose, eliminate, or reduce the activity of different infections.
- nanotechnology
- Klebsiella pneumoniae
- metallic nanoparticles
1. Introductions
Infectious diseases such as pneumonia, diarrhea, tuberculosis, and malaria, are leading causes of death, responsible for 2.2 million, 1.8 million, 1.5 million, and 1.2 million deaths, respectively [1]. More than 95% of the deaths happen in low-income and middle-income countries [2]. The notable increase in drug resistance of infectious agents inhibits efficient therapy for diseases [3]. Klebsiella species are Gram-negative, nonmotile, encapsulated bacteria found in nature, including surface waters, soil, animals, medical devices, and healthcare environments [4]. They were first identified in the late nineteenth century and became known as Friedlander’s bacterium [5]. K. pneumoniae virulence factors play various roles in different K. pneumoniae infections. These virulence agents include fimbriae, siderophores, lipopolysaccharide (LPS), porins, outer membrane proteins (OMPs), iron transport systems, efflux pumps, and genes related to allantoin metabolism [6]. This bacterium can produce biofilms on different levels in the body to escape the host defense. In addition, due to the presence of a polysaccharide capsule that acts as an outer shell, it is significantly protected from phagocytosis by polymorphonuclear granulocytes [7]. In general, K. pneumoniae has long been known as an organism that causes serious primary infections in immunocompromised people. Still, the number of individuals at risk has increased due to the emergence of excessive virulence strains. Even healthy people and persons with adequate immunity are at risk of becoming infected with the bacterium [8]. Klebsiella pneumoniae as an opportunistic pathogen can be easily colonized in human mucous membranes such as the oropharynx and gastrointestinal tract; this colonization appears to be benign [4]. Nevertheless, if this bacterium spreads from the mucosa to other tissues in the body, it can lead to serious and threatening infections such as pneumonia, sepsis, and urinary tract infections (UTIs) [9]. Klebsiella infections cause serious problems in immunocompromised patients, infants, and the elderly. This organism is also one of the bacteria that cause nosocomial infections and community-acquired infections [10]. K. pneumoniae can be divided into hospital-acquired pneumonias (HAPs) and community-acquired pneumonias (CAPs). HAPs are much more common than CAPs, and the underlying cause of 11.8% of HAPs is K. pneumoniae [11]. One of the most serious and dangerous consequences of K. pneumoniae pneumonia and urinary tract infections is bacteremia caused by bacteria entering the bloodstream. K. pneumoniae is the second most deadly bacteria among Gram-negative bacteria as a cause of both community- and hospital-based bacteremia [12]. A K. pneumoniae pathotype known as hypervirulent K. pneumoniae (hvKp) is spreading worldwide. Unlike infections caused by the classical K. pneumoniae (cKp), hvKp causes invasive tissue infections in healthy people in the community and often involves multiple sites [13]. In recent years, the resistance to a wide range of antibiotics of K. pneumoniae has increased significantly. As a result of this increase in antibiotic resistance, infections such as pneumonia and bacteremia are increasingly threatening human health. Simple infections such as urinary tract infections often do not respond well to treatment [14][15][14,15]. The emergence of multidrug-resistant (MDR) bacterial strains is a major threat to patients because, with the spread of this type of antibiotic resistance, most existing treatments fail [16][17][16,17]. Several mechanisms of resistance to antibiotic agents, including the product of ion extended-spectrum β-lactamases (ESBLs) and carbapenemases such as Klebsiella pneumoniae carbapenemases (KPC), over-activation of efflux pumps, and porin modification, have been reported in K. pneumoniae. This is important as the bacterium becomes resistant to all β-lactams, including carbapenems [18]. The emergence of KPC-producing strains has become a significant medical problem. β-lactamases can hydrolyze carbapenem and make this bacterium resistant to a wide range of β-lactam antibiotics. Therefore, treating infections caused by this pathogen has become a considerable challenge [19]. KPCs are frequent in K. pneumoniae but may also be seen in other Enterobacteriaceae types, including Salmonella enterica, Enterobacter species, Escherichia coli, Citrobacter freundii, and Proteus mirabilis [20][21][20,21]. Because of the presence of KPC genes, for example, blaKPC-2, on the plasmid, has a high ability to spread [20][22][20,22]. Due to the lack of appropriate alternative therapies, K. pneumoniae infections created by ESBL-producing strains and carbapenem-resistant strains have a higher mortality rate than non-resistant bacteria [23]. The emergence of MDR K. pneumoniae strains, including colistin-resistant strains, has caused great concern. This type of resistance occurs due to mutations in the mgrB gene, which is stably conserved in Klebsiella populations and by the plasmids carrying mcr-1 and mcr-2 genes [24]. Furthermore, an extensive drug-resistant (XDR) cKp strain, which obtained a segment of a virulence plasmid carrying hvKp, led to a lethal nosocomial outbreak [25]. Today, the search for alternative therapies for antibiotic-resistant K. pneumoniae and other bacteria is vital and essential worldwide [26].
Due to rising antibiotic resistance, K. pneumoniae infection has become a significant health hazard, restricting effective treatments. Therapies reprogramming lung defenses and improving immune response to clear bacteria may help prevent K. pneumoniae infection [27]. Accordingly, in recent years, there has been increased demand for new strategies, pharmaceuticals, and devices to diagnose and treat diseases precisely and competently [28][29][28,29]. Many types of nanoparticles, such as graphene, polymers, vesicles, and green synthesized NPs, have been developed as drug delivery systems in cancer and infectious diseases [30][31][32][30,31,32]. The ability of nanomaterial-based therapeutics and diagnostics to overcome established processes linked to acquired drug resistance makes them intriguing tools for treating difficult-to-treat bacterial infections. Additionally, nanoparticles’ distinctive size and physical characteristics enable them to target biofilms and treat resistant illnesses [33]. Many kinds of nanomaterials have been considered to control infectious diseases effectively [34][35][36][34,35,36]. Nanomedicine has recently been used to increase immune responses to antigens for efficient vaccination, targeted delivery and sustained release of medications, and rapid and reliable disease detection and diagnosis at low cost [37][38][37,38].
2. Nanotechnology-Assisted Approaches for Effective Detection of K. pneumoniae
2.1. Nanoparticle-Assisted Multiple Cross-Displacement Amplification
Numerous isothermal amplifying approaches using nanostructures to amplify nucleic acids in water baths or a simple heating block have been widely reported. The multiple cross displacement amplification (MCDA) test, developed by Wang et al., is a quick and easy method for amplifying nucleic acids at a fixed temperature in under 40 min [39][65]. For example, the detection of K. pneumoniae by label-free MCDA coupled with nanoparticle-based biosensors was developed by Wang et al. [40][66]. The MCDA reaction was carried out for only 30 min at a constant temperature (65 °C), and the amplification findings were directly presented utilizing a lateral flow biosensor (LFB). The results demonstrated that reaction products might be detected in clinical samples with as few as 100 fg and 4.8 CFU of pure K. pneumoniae templates and as few as 480 CFU in 1 mL of spiked clinical specimens. All K. pneumoniae strains tested were positive for label-free MCDA-LFB analysis, while all non–K. pneumoniae strains tested were negative for label-free MCDA-LFB analysis, showing the label-free MCDA-LFB assay’s excellent specificity. The label-free MCDA-LFB test was further enhanced with Antarctic heat-sensitive uracil-DNA-glycosylase (AUDG) to reduce carryover contamination and remove misleading data. As a result, the label-free MCDA-LFB assay combined with the AUDG enzyme proved to be a quick, easy, sensitive, and reliable method for detecting the target pathogen. It also could efficiently avoid carryover contamination, making it a valuable tool for clinical diagnosis, “on-site” identification, and primary quarantine [40][66]. A similar study reported that an MCDA assay for identifying K. pneumoniae could produce positive results after 40 min of isothermal amplification [41][67]. For the quick readouts of MCDA amplification, colorimetric indicators and an Au NP LFB were used. The LOD of this assay was 100 fg per reaction at 65 °C, which was validated by real-time turbidimeters to be the best amplification temperature. For the MCDA assay’s specificity, all 30 clinical-source K. pneumoniae strains were positive, whereas all non–K. pneumoniae strains from 31 different species were negative. The approach’s practicability was used to identify K. pneumoniae strains in sputum samples (24 CFU per reaction) and DNA templates from 100 sputum samples using the MCDA-LFB technique. The MCDA test correctly recognized all of the sputum samples that were positive for K. pneumoniae (30/100) utilizing the culture technique, and its identification sensitivity was greater than that of the polymerase chain reaction (PCR) (25/100). As a result, the MCDA test for K. pneumoniae and the gold nanoparticle LFB is a straightforward, specific, and accurate approach for detecting K. pneumoniae in clinical specimens [41][67]. In addition, in another study, using MCDA and Au NPs LFB, a simple approach for detecting Pseudomonas aeruginosa (P. aeruginosa) was developed [42][68]. This approach performed the reaction in 40 min at an optimal constant temperature (67 °C). An LFB could visually detect the reaction product, obviating the need for specialized equipment. The P. aeruginosa-MCDA-LFB technique was highly specialized for P. aeruginosa and accurately separated it from other infections. It was possible to identify only 10 fg of the genomic DNA template (from pure culture). The assay had the same specificity and sensitivity as the reference (culture-biochemical) approach for detecting P. aeruginosa in clinical sputum samples [42][68]. To discover the mcr−1 gene, Gong et al. employed a similar approach and developed an MCDA linked with the detection of amplified products using an Au NP LFB system [43][69]. The MCDA-LFB test was done at an isothermal temperature (63 °C) for only 30 min during the amplification phase. The reaction products were directly recognized using LFB, which produced results in less than 2 min. The whole test process took about 60 min, from template extraction to the results from evaluation. All of the 16 mcr-1-producing strains were positive, while all of the non-mcr-1 isolates yielded negative findings, demonstrating the analytical sensitivity of this approach. In pure culture, the specificity of the mcr-1-MCDA-LFB test was as low as 600 fg of plasmid DNA per response, while in spiked fecal samples, it was around 4.5 × 103 CFU/mL [43][69]. Because of its low cost, good effectiveness, and real-time identification, the nanopore test has recently been employed for screening biomarkers of diseases [44][70]. For example, Niu et al. separated CRKP from carbapenem-sensitive K. pneumoniae (CSKP) by detecting increased amounts of extracted 16S ribosomal RNA (16S rRNA) from bacterial culture using imipenem, showing that CRKP’s growth was unaffected by the antibiotic [44][70]. The quick and ultra-sensitive identification of 16S rRNA was enabled by specific signals from the single-channel recording of 16S rRNA bound with probes by MspA nanopores. This researchtudy demonstrated that only 4 h of CRKP growth time was required for the nanopore analysis to discriminate between the two proteins. The test takes about 5% of the time of the disk diffusion approach while achieving equivalent precision (See Figure 1) [44][70].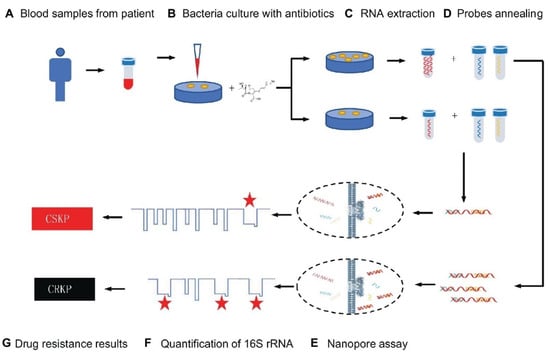
2.2. Nanoparticle-Assisted Loop-Mediated Isothermal Amplification
There have been numerous investigations on the LAMP (loop-mediated isothermal amplification) assay for pneumoniae identification. However, this diagnostic system often produces false positive results due to a high rate of non-specific reactions caused by the formation of hairpin structures, self-dimers, and mismatched hybridization [45][71]. Recent advances have proven that nanomaterials can effectively inhibit undesired amplification and reduce non-specific signals effectively [46][72]. For example, a LAMP-coupled nanoparticle-based LFB assay (LAMP-LFB) was developed for the specific detection of pneumoniae (P-LAMP-LFB) [47][73]. The optimal temperature for this experiment was proven to be 65 °C, and six primers corresponding to the P1 gene of pneumoniae were prepared. Within 2 min, LFB was able to evaluate the amplified outputs accurately. The P-LAMP-LFB test recognized DNA templates of pneumoniae exclusively, with no cross-reactivity with other infections. The LOD of this approach in pure cultures was about 600 fg of the DNA templates, which was in perfect agreement with agarose gel electrophoresis assay and colorimetric indicator identification. This technique was compared to real-time PCR analysis on 209 oropharyngeal swab specimens obtained from children suffering respiratory tract infections. Positive rates of pneumoniae were 47.8% and 31.6%, respectively, using the LAMP-LFB and real-time PCR assays. Compared to the real-time PCR approach, the LAMP-LFB assay exhibited greater selectivity [47][73].2.3. Optical Nanosensors
Colorimetric and fluorescence sensor arrays have generated great interest in recent years because of their capacity to differentiate a wide range of bacteria with a high recognition rate [48][74]. These biosensors rely on the optical characteristics of the sensor surface being altered by the bound analyte, and these changes are subsequently communicated to a detector [49][50][51][75,76,77]. The development of nanotechnology and the application of nanomaterials’ advantages on a biosensor platform allow for an improvement in the biosensors’ sensing parameters. Nanomaterials are employed to great advantage in nanosensors as a label-free detection approach because of their capacity to induce SPR [52][53][78,79]. In this light, a colorimetric nanosensor was developed for selectively identifying bacteria like K. pneumoniae. Four gold nanoparticles (AuNPs) with different surface charges were employed as sensing components. The interactions of AuNPs with microbes resulted in visible hue shifts that could be seen with bare eyes. A total of 15 bacteria exhibited distinct reactions that were effectively distinguished using linear discriminant analysis (LDA). Microorganism combinations may also be determined with ease. This approach is simple, quick (less than 5 s), efficient, and visual, indicating that it could be used for pathogen detection and environmental sensing (See Figure 2) [54][80].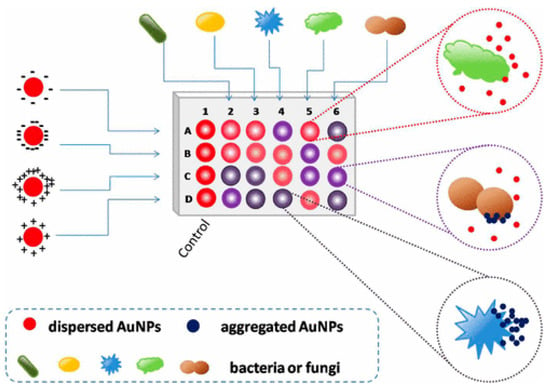
Figure 2. Schematic representation of the colorimetric nanosensor with four types of coated AuNPs. The interactions of microorganisms and AuNPs result in color changes. In the diagram, column 1 represents the blank control, and other columns show various organisms. Rows A to D represent four coated AuNPs, adapted from [54][80], ACS, 2017.
2.4. Cantilevers-Based Nanosensors
As a result of their high responsivity and ease of integration, cantilevers are gaining increasing popularity as new-generation biosensors. Microcantilever-based nanosensors are highly appealing for biomedical purposes because of their fast and real-time analysis, ultrasensitive features, and label-free potential [59][85]. For example, a new array biosensor made up of a microcantilever and gold nanoparticle could detect ultralow quantities of pathogenic bacteria, such as Listeria monocytogenes, Shigella spp., Vibrio parahaemolyticus, Staphylococcus aureus, E. coli O157:H7, Salmonella spp., Listeria monocytogenes, Shigella spp., and many others [60][86]. The process was significantly quicker than traditional methods that need PCR amplification or germi-culturing. Six pairs of ssDNA probes (ssDNA1 + ssDNA2) were designed and verified based on sequence examination of the bacteria’s particular gene. The -S-(CH2)6 parts were attached at the 5′ end of ssDNA1 probes and immobilized as a self-assembled monolayer (SAM) on the surface of cantilevers and finally integrated with Au NPs. In addition, the 6-mercapto-1-hexanol SAM were attached to reference cantilevers to remove interferences from non-specific interactions (Figure 3). Microcantilever array sensors and Au NP platforms can detect quickly and precisely various bacteria with working ranges of 3 to 4 orders of magnitude and LOD of 1–9 cells/mL. There was negligible cross-reaction between the probing of various organisms. This has a promise for quick, combinatorial, and extremely sensitive identification for environmental, clinical, and food items [60][86].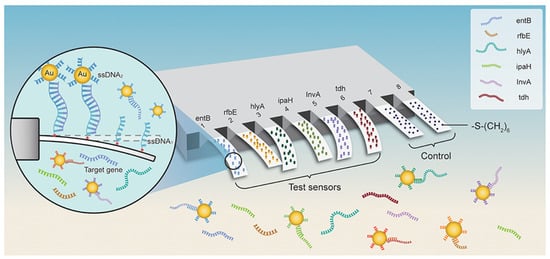
2.5. Electrochemical Nanosensors
Electrochemical nanosensors have potential as a diagnostic tool for people and animals. These sensors can detect pathogens and biomarkers in bodily fluids such as urine and blood [61][87]. For example, the development of a label-free DNA biosensor for the identification of K. pneumoniae that can also diagnose other types of bacterial infections was reported by Zhang et al. [62][88]. In this light, on a glassy carbon electrode, graphene oxide (GO) and indole-5-carboxylic acid (ICA) were successfully deposited, and the resultant ICA/GO (rGO) hybrid film was used to immobilize oligonucleotides on an ssDNA sequence (single-stranded DNA). In optimized conditions, the electrochemical nanosensor showed outstanding performance. A considerable change was seen after hybridizing the target probe with ssDNA under optimal conditions. Differential pulse voltammetry was used to investigate hybridization with three-base mismatched, one-base mismatched, noncomplementary, and complementary DNA targets. With a linear range of 1 × 10−6 to 1 × 10−10 M, the suggested method could identify target DNA as low as 3 × 10−11 M, demonstrating great sensitivity of the nanosensor. The nanosensor showed fast analysis time, devoid of indicators, and had a high degree of specificity. As a result, the proposed nanoplatform can help diagnose K. pneumoniae and other pathogen-related infections [62][88]. Another work presented a glassy carbon electrode (GCE) enhanced with graphene and Au-NPs as a new electrochemical nanosensor for DNA identification. After that, electrochemical impedance spectroscopy, cyclic voltammetry (CV), and scanning electron microscopy (SEM) was used to analyze Au-NPs/Gr/GCE (EIS). In addition, differential pulse voltammetry (DPV) using methylene blue (MB) as the hybridization marker was performed to identify hybridization processes. The sensor’s LOD and dynamic range for the target DNA sequences were 2 × 10−13 mol/L and 1 × 10−12 to 1 × 10−7 mol/L, respectively. In the presence of mismatched and non-complementary DNA sequences, the DNA nanosensor demonstrated remarkable selectivity for recognizing complementary DNA sequences. The findings showed that the Au-NP/Gr nanostructure is a potential substrate for developing high-performance catalytic systems for KPC measurement [63][89].2.6. Biomimetic Nanosensors
Nature is a source of inspiration for solving biological problems. Potentiometric, voltammetric, and impedance spectrum sensors are among the biomimetic nanosensors that mimic the behavior and operation of living organisms by being modified with nanomaterials and specially designed biomimetic materials [64][90]. A new biosensor was designed using graphene and two-photon polymerization to provide an improved biosensor for detecting motile bacteria. Around graphene-based sensing electronics, a cage with a directed micro-architecture was covered that was inspired by venous valves. The designed 3D-mucro architecture enables motile cells to move from the outside of the cage to the center area, leading to an accumulation of bacteria around the core sensing zone, which improves the received signal. The concentrating effect has been shown in cell cultures, ranging from 101 to 109 CFU/mL. Fluorescence evaluation indicated a signal enhancement of 3.38–3.5 fold. Identifying cellar metabolites improves the pH sensor by 2.14–3.08 fold. Electrical tests showed an increase in current of 8.8–26.7 fold. The suggested architecture enables the construction of smart biomedical sensors using bio-inspired 2D materials and 3D printing in a novel manner [65][91]. Table 1 summarizes the nano-based detection method of K. pneumoniae.Table 1. Nano-based detection method of K. pneumoniae.
| Detection Method | Principle | Advantage | Disadvantage | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nanoparticle-assisted multiple cross-displacement amplification | Amplifies the circular DNA template with the use of random primers and DNA polymerase. Within a few hours, the DNA may be amplified over 10,000 times. |
|
| [66] | [92] | ||||
| Optical nanosensors | Utilizes the altered optical characteristics of the nanometric surface of the sensor brought about by the bound analyte, and these altered optical characteristics are then sent to a detector. |
|
| [67] | [93] | ||||
| Cantilever-based nanosensors | A biomolecular interaction produces a change in the mechanical behavior of the transducer (a movement at nanometer scale), which can be measured and analyzed in real time. |
|
| [68] | [94] | ||||
| Electrochemical nanosensors | Transforms the interaction of an analyte with a receptor on the surface of an electrode into a useful analytical signal. |
|
| [69] | [95] |
