You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Mathias Ahii Chia and Version 2 by Amina Yu.
Advances in molecular biology have facilitated the use of polyphasic approaches involving chemotaxonomic, phenotypic, and genotypic data to characterize cyanobacteria. Genotypic diversity through PCR (polymerase chain reaction) amplification of target sequences, cloning, and DNA sequencing of isolated strains and field samples has been poorly described in Africa. The most commonly used genetic markers in Africa include 16S rRNA, PC-IGS, 16S-23S ITS1-L, 16S-23S ITS1-S, rpoB, rpoC1, and 16S-23S ITS. These molecular markers have been employed to understand the taxonomy and phylogeography of cyanobacteria worldwide.
- biological risk
- cyanobacteria
- cyanotoxin
- African continent
1. 16S Ribosomal RNA and 16S–23S rDNA Internal Transcribed Spacer (ITS)
The small subunit ribosomal RNA molecules of ribosomes, which are encoded by the 16S rRNA gene, are necessary for the process of translating genetic instructions into functional cell components via the translation of mRNA to proteins. Because it is a part of all self-replicating systems, ribosomal RNA is very easy to extract, and its slow rate of sequence change over time makes it possible to identify relationships between quite different species [1][41]. The 16S rRNA gene’s sequence was discovered to contain both conserved and changeable sections through a sequence analysis and structural modeling. Variable nucleotides appear at the surface of the ribosome, where it is hypothesized that nucleotide substitutions do not alter ribosome activity, in contrast to evolutionarily conserved nucleotides that appear toward the center of the ribosome at functional regions [2][42].
HIn this rere, 16S rRNA wasview, we retrieved 16S rRNA and ITS1 (Figure 13 and Figure 24) sequences were deposited in the National Center for Biotechnology Information (NCBI) database that had a study or data on African cyanobacteria strains to generate phylogenetic trees using the NJ (neighbor-joining) algorithm: a widely used method for a long time for constructing phylogenetic trees. NJ is a greedy algorithm based on the distance between sequences that endeavors to minimize the sum of all the branch lengths of the resulting tree. It is out of the scope hereinof our paper to cover the limitations or comparisons of methods employed for a DNA sequence analysis. Moreover, all the figures (Figure 13 and Figure 24) contain data from annotated sequences of cyanobacteria blooms and isolated strains in Africa and other continents in the NCBI database. There was a tendency for 16S rRNA sequences of isolated strains from the same country to group independently of the cyanobacterial genera (Figure 13). Furthermore, cyanobacteria samples collected from Kenya and Algeria formed subclusters with cultured Microcystis aeruginosa and uncultured cyanobacteria from Israel and China, respectively (Figure 13). Another subcluster was formed by cyanobacteria isolated from Tunisia, South Africa, Kenya, and Japan.

Figure 13. Neighbor-joining phylogenetic tree of the 16S rDNA gene of bloom samples and isolated cyanobacteria from Africa and other continents. The bootstrap consensus tree was inferred from 1000 replicates, and the evolutionary distances were computed using the maximum composite likelihood method. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1462 bp in the final dataset. Evolutionary analyses were conducted in MEGA X version 11 for macOS.
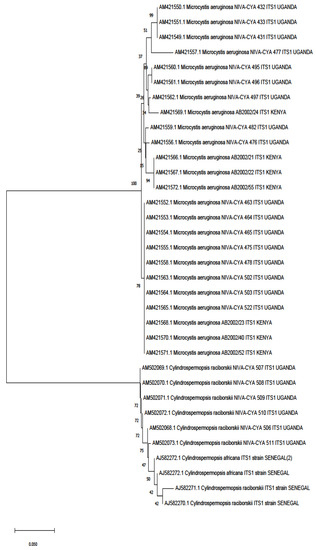
Figure 24. Phylogenetic tree constructed using the neighbor-joining method of the ITS1 sequences’ gene of cyanobacteria detected from studies on the continent. Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. This analysis involved 34 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 539 bp in the final dataset. Evolutionary analyses were conducted in MEGA X version 11.
The combination of molecular markers increases the accuracy of the taxonomic classification of cyanobacteria. Fathalli et al. [3][25] reported using 16S rRNA and rpoC1 molecular markers to characterize four Raphidiopsis raciborskii strains isolated from Tunisia. The rpoC1 gene sequence was used to determine the phylogenetic connections among the strains, which were taxonomically identified using 16S rRNA sequences. The composite molecular markers revealed that Tunisian strains established a unique clade from other African strains included in the phylogenetic analysis. This result demonstrated that rpoC1 sequences can discriminate between close taxa—unfortunately, only a few studies have used this molecular marker in Africa. This strongly corroborates with previous findings showing that the rpoC1 gene sequence is extra selective at the species level compared to the 16S rRNA sequence [4][43]. Furthermore, the maximum likelihood (ML) phylogenetic tree of R. raciborskii worldwide strains inferred from combined sequences of 16S rRNA, ITS-L, ITS-S, and rpoC1 encompassing~2927 bp grouped into three well-supported distinct clusters: the European (I), African/American (II), and Asian/Australian (III) strains [5][31]. The African strains consisted of Tunisian strains. In a review article, Padisák [6][44] postulated that R. raciborskii originated in Africa. Over the years, it has spread to equatorial regions such as Indonesia and Central America, with a second radiation center in Australia, accounting for the more recent cyanobacteria invasion in the tropical, subtropical, and temperate regions. The distinct clustering of African–Australian R. raciborskii strains supports this evidence [7][8][27,45].
Using the Denaturing Gradient Gel Electrophoresis (DGGE) technique coupled with the 16S–23S rDNA internal transcribed spacer (ITS), van Gremberghe et al. [9][46] analyzed the local and regional genetic diversity as well as the structure of Microcystis populations in 30 man-made reservoirs from Tigray (Northern Ethiopia) in wet and dry seasons. Both regional and local Microcystis ITS diversity (number of distinct ITS types in the DGGE profile per sample) were relatively low, with several dense blooms containing only a single ITS type (EG07). In addition, the authors observed a geographic signature and a limited influence of local environmental conditions on the Microcystis population structure, represented by the presence or absence of ITS types in the DGGE profile per sample and their relative abundances. Members of the genus Microcystis are the most investigated cyanobacteria worldwide, and Africa is no exception. Research by Moreira et al. [10][47] suggested that Microcystis aeruginosa originated on the African continent, with further Europe-mediated global dispersal of the species.
2. Amplified Fragment Length Polymorphism
Amplified fragment length polymorphism (AFLP), a PCR-based fingerprinting technique, and SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), a protein profile analysis technique, have been used to characterize cyanobacteria in Africa molecularly. The SDS-PAGE protein profile was used for the first time to analyze the genus Merismopedia in Egypt (North Africa) [11][48]. Oberholster et al. [12][38] employed AFLP fingerprinting to highlight the differences among 23 strains of geographically unrelated Microcystis isolated from South Africa, and the UPGMA (unweighted pair group method with arithmetic mean) hierarchical clustering method revealed four groups. The NIES strains of Japanese origin clustered in Group 1; the European strains clustered in Group 2; the South African strains (northern SA) formed the third distinct group; and the strains collected from the central and southern regions of SA clustered together with the US strains in Group 4. Their results revealed extensive evidence of the applicability of AFLP in cyanobacterial characterization, validating its discriminative power towards the differentiation of unrelated Microcystis strains of the same species (M. aeruginosa), as well as highlighting the potential of genomic restriction fingerprinting in evolutionary studies.
3. Omics—Metagenomics for Characterization of Cyanobacteria in Africa
Metagenomic studies of cyanobacteria are scarcely conducted in Africa. Using multilocus 454-amplicons sequencing, Dadheech et al. [13][32] used metagenomics to investigate the cyanobacterial community structure of Lake Bogoria, Kenya. A high degree of endemism was found for Oscillatoriales phylotypes (Leptolyngbya, Spirulina, Oscillatoria-like, and Planktothricoides) in the Bogoria hot springs. Different clades of Synechococcus represented Chroococcales. The pelagic zone and the sediments were inhabited by only a few taxa, dominated by Arthrospira and Anabaenopsis. Arthrospira, the main food for Lesser Flamingos, was detected in all three habitats (hot springs, pelagic zone, and sediment), indicating its resilience and critical role as a primary producer. We Slook forward to seeing additional metagenomics studies in Africa is expected, because taxonomic classification using this approach enables the establishment of the exact origin of species and cataloging or classifying various cyanobacterial groups inhabiting unique environments in Africa.
