ANTEA (GI) is a gene involved in multiple biological functions, which have been analysed and are partially conserved in a series of mono- and dicotyledonous plant species. The identified biological functions include control over the circadian rhythm, light signalling, cold tolerance, hormone signalling and photoperiodic flowering. The latter function is a central role of GI, as it involves a multitude of pathways, both dependent and independent of the gene CONSTANS(CO), as well as on the basis of interaction with miRNA. The complexity of the gene function of GI increases due to the existence of paralogs showing changes in genome structure as well as incidences of sub- and neofunctionalization. We present an updated report of the biological function of GI, integrating late insights into its role in floral initiation, flower development and volatile flower production.
- Gene ontology
- molecular function
- cellular localisation
- biological function
- circadian clock
- flowering time
- flower development
- floral scent
1. Introduction
GIGANTEA (GI) is a plant-specific nuclear protein, identified for the first time in Arabidopsis thaliana as a late flowering mutant[1]. Although six decades have passed since its discovery, its precise molecular function has not been completely elucidated. Only at the end of the XXth century was it possible to obtain information describing its chromosomal organization[2]. The mapping identified the genomic locus on chromosome 1, consisting of 14 exons and encoding for a protein of 1,173 amino acids[2][3]. The GI gene, which appeared early in land plants, is present in a single copy in most plants, such as Arabidopsis or rice[4], while in Solanaceae it is found in two or three copies[5]. Evolutionary phylogenetic analysis of the gene has shown that GI in Petunia and in general in Solanaceae, is grouped separately from the clade of Brassicaceae, Rosaceae and Fabaceae. This indicates an evolutionary departure that appears to be specific to plant families. Further gene duplications have been found in the subclades of tomato, Nicotiana benthamiana and Petunia inflata [5].
Recent studies carried out in Petunia[6] support the hypothesis that the structural evolution of the main circadian clock genes such as PhGI, occurred through changes in the number of paralogues via gene duplications. Further changes have occurred in the gene structure and in the coding region, including both deletions and substitutions, depending on the species and paralog.
This review gives an overview of the molecular functions, cellular localisation and biological functions of GI paralogs, described in model plants as well as crops of agronomic importance.
2. The Cellular Localisation of Gigantea
The cellular localisation of GI in Arabidopsis was identified by constitutively expressing GI-GFP both transiently in protoplasts and transgenic plants. Fluorescent microscopy analysis revealed that GI protein is predominantly present in the nucleus of some cell types, forming nuclear bodies[7][8]. Importantly, GI proteins lack a nuclear localisation signal. To better understand the nature of these formations, specific sub-nuclear marker-proteins of different compartments were used, directed specifically to nucleoli, spliceosomes, heterochromatin beams and Cajal bodies. GI was not localized in any of the aforementioned nuclear compartments, showing that GI does not play any role in the processes of protein degradation, pre-mRNA splicing and biogenesis of rRNA and snRNP. The evening complex gene EARLY FLOWERING 4 (ELF4) was found to sequester GI from the nucleoplasm, where it binds to the CO promoter, to discrete nuclear bodies [8]. This process is induced under long days in Arabidopsis. ELF4 protein synthesis oscillates during the day and consequently does the formation of GI nuclear bodies, peaking at night, and it is proposed that GI can then be released in the morning, avoiding the necessity for de novo GI synthesis[8].
GI is also localized in the cytosol where it stabilizes the F-box protein ZEITLUPE (ZTL) through heterodimerization. Under blue light, GI binds to the LOV (light, oxygen or voltage sensing) domain of ZTL, thus ensuring a robust and accurate oscillation of its target proteins[9]. It was proposed that sequestration of GI by ZTL to the cytosol, regulates the nuclear pool of GI and controls thereby the distribution of GI between nucleus and cytosol as well as its protection from degradation[10]. Degradation in the nucleus coincides with the high accumulation of CONSTITUTIVE PHOTOMORPHOGENIC 1(COP1) and clock-associated protein ELF3. COP1 is an E3 ubiquitin-ligase, and ELF3 allows COP1 to interact with GI leading to GI degradation under short day conditions. Thus, the destabilisation of GI through COP1 and ELF3 plays an important role in the modulation of circadian rhythms and regulation of flowering transition in Arabidopsis[11].
3. The Molecular Functions of Gigantea
3.1. Gigantea Coordinates Light Signalling, Protein Degradation and Transcription of the Circadian Clock
The most recent clock model, based on Arabidopsis, describes the endogenous clock as an intricate system of negative autoregulatory feedback loops interacting with each other via transcriptional and post-translational activation and repression[12] (Figure 1). Two MYB domain transcription factors, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) form the central loop together with PSEUDO-RESPONSE REGULATOR 1 (PRR1), also known as TIMING OF CAB EXPRESSION 1 (TOC1). The morning loop is formed by PSEUDO-RESPONSE REGULATORs (PRRs) 9,7 and 5[13]. The evening loop is composed of EARLY FLOWERING 3 and 4 (ELF3 and ELF4) and LUX ARHYTHMO (LUX), together with ZEITLUPE (ZTL)[14]. ELF genes and ZTL control light input signals to the clock and the ability of plants to distinguish between different day lengths[2][9][15]. GI interacts with several of these clock genes, ensuring expression peaks, period length and amplitude of the different clock genes at specific times of the day. In Arabidopsis, GI transcript levels are controlled by the circadian clock with peak transcript levels at 8–10 h after dawn[2]. In Petunia, a lack of robust circadian rhythmicity under continuous darkness for PhGI1 hints to the necessity of correct light signalling for oscillation[16].
One molecular function within the circadian oscillator complex consists of the binding of GI protein to ZEITLUPE (ZTL), a protein codified by a gene belonging to the evening complex. ZTL receives light inputs through its LIGHT, OXYGEN, VOLTAGE (LOV) domain, sensing blue-light, but it also has an F-box, targeting protein for degradation. ZTL degrades the central clock protein TOC1[14]. GI is crucial for stabilizing and maintaining the oscillations of ZTL blue-light photoreceptor through direct protein protein interaction. In this connection, GI interacts with the blue light-sensitive domain of ZTL, stabilising it post-translationally under blue light[9][17]. In Arabidopsis, the expression of ZTL is uniform during 24 hours, while protein levels oscillate[18]. In contrast, transcription of PhCHL, the Petunia ortholog of ZTL, is characterized by a significant rise at midday[16][19]. There are two paralogs of GI in Petunia x hybrida, PhGI1 and PhGI2. The Silencing of PhGI1 causes a significant prolongation of the rhythmic period of expression of PhCHL[16]. This suggests that coordination of PhCHL by PhGI does not only occur at the post‐translational level, but that PhGI also coordinates the timing of expression of PhCHL during the day. However, it is not clear yet if this is a direct effect or happens as a result of modified levels of proteins involved in transcription such as PhTOC1.
According to the theory of the repression feedback circuits, the morning elements LHY and CCA1 heterodimerize and repress the expression of TOC1 and the evening complex members as GI, LUX, ELF3, ELF4. PRR9, PRR7 and PRR5, which in turn, when expressed, repress the expression of CCA1 and LHY. In the evening, TOC1 represses all of the previously expressed components. The molecular interaction between GI and ZTL on one side and ZTL and TOC1 on the other side, predicts changes in the expression pattern of clock genes affected by TOC1 protein levels. Indeed, experiments carried out by Fowler et al. (1999) in Arabidopsis demonstrated that under both long days (LDs) and short days (SDs), a mutation in the GI locus affects the CCA1 and LHY gene expression. On the other hand, over-expression or mutations of CCA1 and LHY disrupt the GI expression. In agreement with these results, the double mutant of LHY and CCA1 shows an overabundance of the transcription of GI [7]. These results led to the conclusion that GI operates in a feedback loop for adjusting and maintaining the length of the clock period. The involvement of GI in circadian clock control was confirmed in rice, were OsGI controls amplitude and circadian rhythm phases of global gene expression under natural field conditions[4].
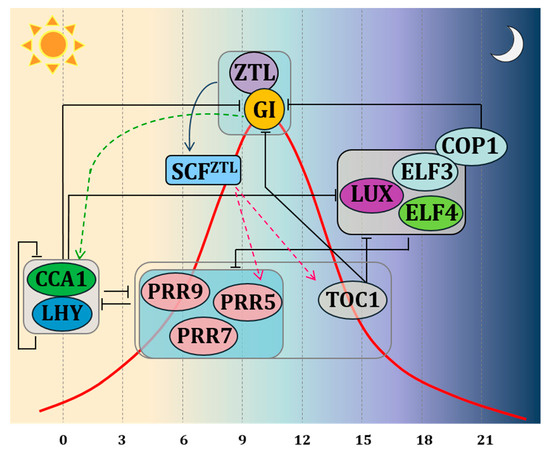
Figure 1. Schematic representation of the clock model indicating interactions by autoregulatory feedback loops via transcriptional and post-translational activation and repression. The clock components are depicted according to their peak-time expression from left to right. Black bars indicate repression. Green arrowheads lines indicate activation of transcription. Fucsia arrowheads lines indicate proteasomal degradation. The red curve represents the expression of GI during the day. TOC1 is protected during the day from ZTL-induced proteasome degradation thanks to the direct protein-protein interaction of GI with ZTL. During the evening, ELF3 promotes the proteasome degradation of GI through its interaction with COP1, thus promoting the ZTL-dependent TOC degradation. Boxes indicate sets of genes acting in coordination. The PRR box: PRR 5,7,9—these proteins are sequentially expressed during the day, and all repress LHY and CCA1; GI-ZTL box—protein-protein interaction leads to ZTL maturation; ELF3-ELF4-LUX complex box—protein complex peaking at dusk.
3.2. Flowering Time and Photoperiod Related Molecular Functions
In Arabidopsis, photoperiod, causing the transition from the vegetative phase to flowering, is regulated by the diurnal expression of CONSTANS (CO)[20][21]. It has been shown that during long days (LD), light stabilises CO protein. In Arabidopsis, the expression of CO coinciding with the period of light leads to the activation of FLOWERING LOCUS T (FT) gene. On the other hand, under short day (SD) conditions, the peak expression of CO occurs after sunset because the CO protein is not sufficiently stabilized by light[22]. The transcription of CO is repressed during sunrise, due to the activity of the CYCLING DOF FACTOR 1 (CDF1) transcriptional repressor bound to the CO promoter.
GIGANTEA plays a key role in this flowering regulation pathway. Figure 2 illustrates the involvement of GI in flowering time control in Arabidopsis as well as in other model plants. Although several paralogs of GI exist in different species, i.e., maize, thus far, the existing models on flowering time control represent the function of only one of the paralogs, and further research may elucidate further functions of GI in other plants. In Arabidopsis, an enzymatic complex is formed thanks to a direct protein-protein interaction between GI and FLAVIN-BINDING, KELCH REPEAT, F BOX protein 1 (FKF1), through its FKF1 LOV (Light, Oxygen or Voltage) domain[23]. This complex mediates the degradation of CYCLING DOF FACTOR 1 (CDF1), a main CO repressor. This complex is strictly dependent on light, being the expression of GI under the control of the circadian clock. Therefore, towards the middle of the day, when the accumulation of GI along with FKF1 reaches the peak, the DOF-degradation complex is repressed, leading to an increase in the transcription of CO and, therefore, to the transcription of FLOWERING LOCUS T (FT)[23][24]. This does not happen in short days in Arabidopsis, as the GI accumulation peak occurs about three hours earlier than that of FKF1. This prevents the formation of the DOF-degradation complex and the consequently reduced abundance of the CO transcript.
Another light-dependent control of flowering through GIGANTEA involves SPINDLY (SPY). GI inhibits SPY in a light dependant manner. At the same time, SPY represses the expression of CO and FT, as observed in spy-4 plants, where the late-flowering phenotype of gi-2 plants was abolished due to a partial suppression of abundance reduction in CO and FT RNA[25]. Furthermore, SPY also affects flowering by inhibiting GA signaling, required for flower promotion[26].
Additionally, GI can regulate the expression of FT independently of CO. It inhibits the FT transcriptional repressors such as SHORT VEGETATIVE PHASE (SVP), TEMPRANILLO 1 (TEM1) and TEMPRANILLO 2 (TEM2). GI alters their stability or neutralises their repressor effect by blocking their access to the FT promoter region. Likewise, GI binds their specific target regions on the FT promoter[27], thus affecting the abundance of the FT transcripts.
GI regulates FT expression and photoperiodic flowering, independent of CO, via its interaction with a microRNA. The miRNA172 inhibits the expression of the main transcriptional repressors of FT, TARGET OF EAT 1 (TOE1) and APETALA 2 (AP2). It was demonstrated that miRNA172 processing is positively regulated in the presence of GI protein through an unclear molecular interaction[28]. Growth of plants under natural conditions, i.e., with photo and thermoperiod, shows that it has a major impact in the coordination of FT, suggesting that GI may have a role in temperature and light coordination[29].
Many GIGANTEA orthologs have been described in the last decades, in a wide range of plant species, from gymnosperms to mono y dicotyledon angiosperms. Many of these have similar expression patterns to those of Arabidopsis GI, suggesting broad conservation of the photoperiodic flowering regulation mechanisms (Figure 2)[30][31][32][33][34][35][36]. Indeed, the GI ortholog of soybean, GmGIa, controls flowering time by inducing the expression of the soybean florigen gene ortholog GmFT2a[37] . Among three haplotypes (H1, H2, H3) of GmGIa, H1 rescues the late-flowering phenotype of gi-2 in Arabidopsis, while H2 and H3 delay flowering in transgenic Arabidopsis with a wild type background. This diversification was proposed to be related to flowering time adaptation during soybean domestication[38]. Similar to Arabidopsis, GmGIa also positively regulates gma-miR172a, which in turn represses Glyma03g33470, leading to upregulation of FT, AP2 and LFY and early flowering[39].
Wheat is a long-day (LD) plant and one GIa ortholog TaGI1, has been described. It shares 63% identity with AtGI and a superior homology to GI from grasses such as rice and barley[30]. Overexpression of TaGI1 alters flowering time in wheat, resulting in early flowering both under LD and SD[30]. In barley, an upregulation of HvGI observed in HvELF3 mutants results in an early flowering phenotype[40].
Conservation of GI function upon flowering time is also observed in short-day (SD) plants. The comparison of the sequences between Arabidopsis GI and its ortholog in Oryza sativa GI (OsGI) reveals 67% of identity as well as conservation of its nuclear localisation[41][42]. In rice, a quantitative SD plant, over-expression of the GIGANTEA gene (OsGI) under LD and SD leads to a delay in flowering time with a more pronounced effect under SD [43]. Rice overexpressing OsGI and under LD condition, show increased expression of the CO ortholog Hd1, while the expression of Hd3a, the FT ortholog, was reduced. Thus, a reverse regulation of Hd3a by Hd1 occurs in rice under LD conditions. Opposite to Arabidopsis, OsGI acts as a flowering time suppressor[43][44][45]. However, under SD conditions Hd1 promotes heading coinciding with increased transcript levels of Hd3a. This suggests that the regulatory network of the key flowering genes is conserved among rice and Arabidopsis[44]. Likewise, in the SD and C4 plant maize, which has two circadian regulated GI paralogs, Zmgi1 mutants flower earlier than non-mutant plants in LD photoperiods, which is the opposite to the late-flowering gi mutants from Arabidopsis, but not in SD photoperiods [36]. The flowering time control mechanism also involves FT-like floral activator gene Centroradialis8 (ZCN8) and the CONSTANS-like flowering regulatory gene Constans of Zea mays1 (CONZ1), which are both upregulated in the Zmgi1 mutant, confirming the existence of the conserved network of the three key regulatory genes Zmgi1, CONZ1, ZCN8 in maize, but gene interaction is photoperiod dependent and differs whether the species is a LD or SD plant.
Petunia x hybrida has two GI paralogs, PhGI1 and PhGI2. Downregulation of PhGI1 by interference RNA does not induce late flowering, but a flowering time effect of PhGI2 cannot be ruled out[16]. In the perennial poplar, three GI-like genes PagGIa, PagGIb and PagGIc were identified and overexpressed in Arabidopsis wild type ecotype Columbia-0 (Col-0), leading to early flowering[46]. Recent work in white lupin has identified LaGI as a major QTL involved in flowering time[47]. As the single copy gene MpGI coordinates phase transition into reproductive development in the basal angiosperm Marchantia polymorpha[48]. Indeed, loss of MpGI causes complete loss of long-day dependent phase transition. We can conclude that that GI plays a key role in flowering time in a variety of species.
Figure 2. Models of GIGANTEA dependent flowering time regulation in (a) Arabidopsis, (b) soybean, (c) rice (d) and maize under long day condition. Regulation of flowering through GI differs between LD and SD plants. In contrast to LD plant Arabidopsis, OsGI inhibits flowering in rice, an SD plant, because the FT ortholog Hd3a is repressed by Hd1, the CO ortholog.
3.3. Light Signalling Related Molecular Functions
Photoreceptors, such as phytochromes, phototropins, cryptochromes and UV-light photoreceptors, control light-induced plant development by the integration of light cues from the environment, such as quality, intensity and duration. Specific wavelength inputs are transformed into physiological signals, a process called photomorphogenesis[49][50]. Cryptochromes and phototropins absorb mainly the blue spectrum (B, λ = 400–499 nm) and are involved in the regulation of flowering time, inhibition of hypocotyl growth and phototropism[51]. Five members of phytochromes (Phy) exist in Arabidopsis, from PhyA to PhyE[42][52]. They act as red (R, λ = 660 nm; PhyB-E) and far-red (FR, λ = 730 nm; PhyA) photoreceptors. PhyA mediates two distinct photobiological responses: The very-low-fluence response (VLFR) and the high-irradiance response (HIR)[49] [49]. Arabidopsis gi-mutants, grown under saturated R light condition, have shown an elongated hypocotyl compared to the wild type and little or no change in responsiveness to continuous FR light[42], indicating that GI appears to be a positive regulator of PhyB signalling during seedling de-etiolation[25]. Considering also that neither the genes nor the abundance of PhyA-B proteins are affected in gi-mutants, it appears that GI works downstream of PhyA-B, after their migration to the nucleus in response to light, where GI is constitutively localised. Additional data regarding gi-mutants grown in FR light condition revealed low VLFR levels, deficient cotyledon unfolding and a low seed germination index. These phenotypes were rescued through GI over-expression, demonstrating that GI plays a role in the PhyA signalling[53]. The gi-mutant seedlings also exhibited a long hypocotyl phenotype when grown under blue light, proving to have a role also in the signalling of blue light[50].
3.4. Hormone Signalling and Stress-Response Related Molecular Functions
One of the plant hormones, whose signalling interacts with GI, is gibberelins. In Arabidopsis, GI affects the growth of the hypocotyl through the interaction with SPINDLY (SPY), a gene involved in the regulation of gibberellin signalling. SPY is, in fact, a negative regulator of gibberellin signalling and an inhibitor of hypocotyl lengthening[25][26]. GI interacts with a protein-protein interaction domain of SPY, consisting of 10 tetratricopeptide repeats (TPRs). Three pathways are controlled through SPY-GI interaction: Flowering, circadian cotyledon movements, and hypocotyl elongation. In the case of the flowering pathway, the reduction of CO and FT RNA abundance in gi plants is partially suppressed in the spy mutant.
GI also interacts with gibberellin signalisation through its interaction with REPRESSOR OF ga1-3 (RGA), a DELLA protein. Under LD, warmer temperatures induce accumulation of GI, which functions as thermostabiliser of RGA, which leads to the attenuation of thermomorphogenesis mediated through PHYTOCHROME INTERACTING FACTOR 4 (PIF4). Under SD and lower GI levels, RGA is degraded through the gibberellic acid-mediated ubiquitination-proteasome pathway[54][55]. An effect of warm temperatures on GI accumulation was concluded from observations on GI-deficient gi-2 mutants, which showed hypocotyl overgrowth at 28 °C but not at 23 °C. Overgrowth at warmer temperatures was related to an increased auxin action, as both YUCCA8 (YUC8), an auxin biosynthetic enzyme, and SMALL AUXIN UPREGULATED RNA 22 (SAUR22), an auxin-responsive protein, were upregulated in gi-2 mutants at warmer temperatures[54].
The Arabidopsis mutant abz126 bears a T-DNA insertion into the ninth exon of the GIGANTEA (GI) gene, resulting in a loss-of-function of GI, leading to long petioles, tall plant height, many rosette leaves and late flowering, as well as insensitivity to paclobutrazol and brassinolide, indicating an association of GI with brassinosteroid hormone signalling[56].
Interaction between GI and stress response exists concerning tolerance to salt. Under normal conditions, GI interacts with the protein kinase SALT OVERLY SENSITIVE 2 (SOS2), thus preventing its interaction with the Na+/H+ anti-porter SOS1 to promote Na2+ export and salt tolerance. Under saline conditions, GI is degraded by the proteasome. Due to GI degradation under salt stress, SOS2 interacts with SOS3 and this protein kinase complex then activates SOS1, salt tolerance and retardation of flowering[57][58].
Opposite to salt stress, drought stress accelerates flowering time, and GI was found to be a key component mediating drought response in Arabidopsis. One proposed mode of action predicts that GI may regulate chromatin accessibility and/or interfere with repressor activity at the florigen promoters in a plant stress hormone abscisic acid (ABA) dependent manner[59].
GI also has molecular functions involved in freezing tolerance. GI activates the expression of CO and FT, the key floral regulators, by facilitating the degradation of a family of CDF1, which acts as transcriptional repressors. In Arabidopsis and tomato, in a gi-mutant background, increased stability and accumulation of CDF proteins and higher transcript levels of stress‐responsive genes, including COR15a, RD29A and ERD1, were observed as well as an increased level of protection against cold stress[60][61]. Therefore, proposed that, in addition to flowering, the regulation of CDFs through GI, influences responses to freezing temperatures[61].
4. Biological Functions of Gigantea
The described molecular interactions of GI as well as its cellular location, result in a wide range of biological functions analysed in Arabidopsis. Part of these functions were found to be conserved in many other plant species, both monocots and dicots. Biological functions include circadian clock regulation, light signalling, flowering time control, which were already described in detail under molecular functions, as well as chlorophyll accumulation, sugar metabolism, stress tolerance, vegetative growth, flower development and floral scent emission described below.
4.1. Chlorophyll Accumulation
The analysis of different Arabidopsis gi-mutants hint to a GI function in chloroplast biogenesis and chlorophyll accumulation—factors essential for photosynthetic efficiency and therefore crop productivity. The Arabidopsis gi-2 mutant is characterized by a reduced sensitivity to lincomycin, a chloroplast biogenesis inhibitor, and the mutant maintains high levels of photosynthetic proteins. In contrast, GI-overexpressing plants have variegated leaves, reduced photosynthetic protein levels and are sensitive to lincomycin[62]. A series of Arabidopsis mutants in Ler background (gi-3,4,5) showed a significantly higher chlorophyll content in seedlings[62]. RNA interference of one GI paralogue in Petunia x hybrida, PhGI1, leads to leaves with a greener appearance in the denser apical region coinciding with a progressive increase in chlorophyll content compared to the wild type[63] (Figure 3a–c). These results thus point to a general role of GI in chlorophyll homeostasis.
4.2. Sugar and Starch Metabolism
Sugars function in plants as a source of energy and as a signal, among others, in the circadian regulation of flowering[64]. The Arabidopsis circadian system was described to be sensitive to sucrose in the dark, indicating feedback between metabolism and circadian clock, and it was predicted that GI is required for the full response of the circadian clock to sucrose[65]. Indeed, GI protein is stabilized by sucrose in the night, and this mechanism requires interaction with ZTL[66].
Furthermore, the Arabidopsis gi-3 mutant is characterized by an enhanced freezing sensitivity, and this was related to a reduction in soluble sugar content in leaves. Transcript levels of the cold-responsive gene RD29A and abscisic acid-responsive gene RAB18 were not affected in this mutant, indicating a direct connection between GI gene function and sucrose metabolism[67]. In contrast to this observation, field-grown rice plants carrying a null mutation in the rice homolog OsGI showed a significantly increased sucrose and starch content in the leaves at most time point[4]. Increased starch content was also observed in gi-1, 2 and 3 alleles in Arabidopsis [68]. A recent proteomic analysis of interactors of GI has identified TREHALOSE-6-PHOSPHATASE SYNTHASE 8 (TPS8) as a direct interactor[69]. This indicates a possible direct link between GI and sugar metabolism.
4.3. Stress Tolerance
Plants cope with environmental stresses by activating a series of specific metabolic pathways. Regarding low-temperature stress, they must avoid freezing injury that can occur in both intracellular and extracellular compartments, prevent chilling wounds and cell injuries that could cause tissue death. The response to cold temperatures implies alterations in the expression of genes, followed by increases in the levels of metabolites, including those known to have protective effects against the damaging effects of cold stress [70]. Plants have the ability to adapt to low temperatures by increasing their freezing tolerance through the gradual exposure to low but non-freezing temperatures, a process identified as cold acclimation[71]. Plant resistance to cold appears to be organ-specific[72]. This process is characterised by complex biochemical and physiological changes, including gene expression[73][74], enzyme activities[75], lipid membrane composition[76] and leaf ultrastructure modification[77]. Chilling injuries cause the collapse of deep cell layers and disruption of antioxidant activity amongst other effects[78]. The biological function of GI concerning freezing tolerance results from its interaction with CDF proteins. GI facilitates the degradation of CDF proteins. Arabidopsis gi-mutants accumulate CDFs accompanied by higher transcript levels of stress‐responsive genes and increased cold stress[60][61]. Additionally, enhanced freezing sensitivity in gi-mutants is also related to a reduction in soluble sugar content[67]. Another direct biological function of GI is related to salt stress. This stress leads to proteasomal degradation of GI[57][58]. GI function in salt tolerance was found to be conserved in poplar, as overexpressing PagGIs in wild type (WT) Arabidopsis induces salt-sensitivity.
4.4. Vegetative Growth
Observations on plant development in Arabidopsis indicate that GI is a negative regulator of vegetative growth as can be inferred from its name as a mutant. Loss of function of GI effects hypocotyl growth, but it also results in long petioles, tall plant height and many rosette leaves[68]. The latter is proposed to be related to gibberellin signalling, as SPINDLY (SPY) protein, a negative regulator of gibberellin signalling in Arabidopsis and an inhibitor of hypocotyl elongation, interacts with GI protein[25]. Similarly, downregulation of the GI1 paralog in Petunia by RNA interference results in bigger leaves (Figure 3a–c), an increased basal internode length, and an increased number of axillary meristems. Middle and apical internodes are reduced, indicating the existence of an acropetal gradient with opposite effects during the early stages of development and middle to late stages (Figure 3d,g). Similarly, downregulation of PagGIs in poplar by RNA interference leads to vigorous growth, and higher biomass[46].
4.5. Flower Development and Floral Scent Emission
A new role of GI in the development of reproductive organs has recently been described in Petunia x hybrida. Plants with loss of GI1 function, grown under LD conditions, show a reduction in the total number of floral buds and smaller flowers than wild type. Flowers show a significant reduction in the corolla diameter as well as in the tube length (Figure 3e). There is an additional floral bud at the bifurcation point, where a given shoot that terminates in a flower and a new sympodial shoot separate (Figure 3f). The aborted flowers clearly show early development of carpel and stamen tissues, indicating that the senescence process occurs after the activation of the floral organ identity genes. These results indicate that PhGI1 is a repressor of early flower senescence, a biological function that had not been described before. The significant reduction in the total number of flower buds also indicates an upstream effect related to the flower-meristem-identity genes PETUNIA FLOWERING GENE (PFG) and ALF (ABERRANT LEAF AND FLOWER)[79][80].
Among the main strategies that plants use to ensure entomophilic reproduction is the emission of floral scents in the form of volatile organic compounds (VOCs). Plant scents are mixtures of volatile lipophilic molecules of benzenoids/phenylpropanoids and fatty acid derivatives with low molecular weight and high vapor pressure at ambient temperature, synthesized in all plant organs, from roots to flowers[81][82]. The plant VOCs are classified into different classes according to their biosynthetic origins, such as: Terpenoids, benzenoids/phenylpropanoids, fatty acid derivatives, and amino acid derivatives as well as few other genus-specific species[83]. In vegetative organs, VOCs are part of the plant’s defence system and are mainly synthesised in glandular trichomes[84], single specialized cells[85] or tubes [86] from which they can sprout out in case of breakage. In many Angiosperms, the quantity and composition of the VOCs can fluctuate during the day, mainly in relation to the age of the flower. This rhythmic release of scent is also closely related to the flower hormonal regulation, circadian clock, flower and plant development, nutrient availability, temperature, humidity and general environmental conditions[87][88].
In recent decades, many studies on the production and regulation processes of VOCs in plants have been carried out, but few genes have been characterized involved in the regulation of scent production. The studies conducted by Verdonk et al., [89] in Petunia x hybrida, have identified a R2R3 MYB-type gene named ODORANT1 (ODO1) which controls the synthesis of the precursor Phe in the shikimate pathway. The Petunia x hybrida CHANEL (PhCHL) gene, the ortholog of Arabidopsis thaliana ZTL, has been shown previously to play an important role in regulating both the timing and the quantity of volatile emissions[19]. Methyl benzoate is the dominant floral VOC in Petunia hybrida. Recent studies on PhGI1 in Petunia x hybrida have revealed thus far unreported functions of this pleiotropic gene concerning the control over VOC emission. PhGI1 loss of function plants exhibited a 20% reduction in the total emission of VOCs during 24 hours. The circadian volatile emission pattern in the silenced lines remained unchanged. However, an important twist in the scent profile was observed, compared to the wild type and non-transgenic siblings with changes in the relative abundance of the trans-cinnamic acid derivatives benzyl alcohol, ethyl benzoate, benzyl benzoate and isoeugenol, indicating involvement of GI in the phenylalanine emission pathway, interfering in the rhythmic regulation of the VOC biosynthesis and their daily emission profile. Interestingly, a null mutation in the rice homolog OsGI, affects the production of several metabolites in the phenylpropanoid pathway, which was significantly increased, whereas the pool of Phe, the major chemical precursor in the pathway of phenylpropanoids, was significantly decreased[4]. These convergent results suggest the role of GI in the control of Phe and phenylpropanoids in plants.
Figure 3. Phenotypic characteristics in Petunia x hybrida plants with loss of GI1 function. Apical (a), median (b) and basal (c) leaves of wild type Petunia (left) compared to iRNA: PhGI1 leaves (right). (d) Vegetative growth architecture of Petunia wild type (left) compared to the loss of PhGI1 function (right). Petunia inflorescence appearance (e) of wild type (left) and PhGI1 silenced line (right). Abortive flower (f) and (g) schematic representation of variations in internode length and number of axillary shoot meristems (indicated by the green colour) between wild type plants (left) and iRNA: PhGI1 plants (right).
References
- György P. Rédei; Supervital Mutants of Arabidopsis. Genetics 1962, 47, 443-460.
- Sarah Fowler; Karen Lee; Hitoshi Onouchi; Alon Samach; Kim Richardson; Bret Morris; George Coupland; Jo Putterill; GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. The EMBO Journal 1999, 18, 4679-4688, 10.1093/emboj/18.17.4679.
- D. H. Park; Control of Circadian Rhythms and Photoperiodic Flowering by the Arabidopsis GIGANTEA Gene. Science 1999, 285, 1579-1582, 10.1126/science.285.5433.1579.
- Takeshi Izawa; Motohiro Mihara; Yuji Suzuki; Meenu Gupta; Hironori Itoh; Atsushi J. Nagano; Ritsuko Motoyama; Yuji Sawada; Masahiro Yano; Masami Yokota Hirai; et al.Amane MakinoYoshiaki Nagamura Os-GIGANTEA Confers Robust Diurnal Rhythms on the Global Transcriptome of Rice in the Field. The Plant Cell 2011, 23, 1741-1755, 10.1105/tpc.111.083238.
- Aureliano Bombarely; Michel Moser; Avichai Amrad; Manzhu Bao; Laure Bapaume; Cornelius S. Barry; Mattijs Bliek; Maaike R. Boersma; Lorenzo Borghi; Rémy Bruggmann; et al.Marcel BucherNunzio D'agostinoKevin M. DaviesUwe DruegeNatalia DudarevaMarcos Egea-CortinesMassimo DelledonneNoe Fernandez-PozoPhilipp FrankenLaurie GrandontJ. S. Heslop-HarrisonJennifer HintzscheMitrick JohnsRonald KoesXiaodan LvEric LyonsDiwa MallaEnrico MartinoiaNeil S. MattsonPatrice MorelLukas A. MuellerJoëlle MuhlemannEva NouriValentina PasseriMario PezzottiQinzhou QiDidier ReinhardtMelanie RichKatja R. Richert-PöggelerTim P. RobbinsMichael C. SchatzM. Eric SchranzRobert C. SchuurinkTrude SchwarzacherKees SpeltHaibao TangSusan L. UrbanusMichiel VandenbusscheKitty VijverbergGonzalo H. VillarinoRyan M. WarnerJulia WeissZhen YueJan ZethofFrancesca QuattrocchioThomas L. SimsCris Kuhlemeier Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nature Plants 2016, 2, 16074, 10.1038/nplants.2016.74.
- Marta I. Terry; Marta Carrera-Alesina; Julia Weiss; Marcos Egea-Cortines; Transcriptional Structure of Petunia Clock in Leaves and Petals. Genes 2019, 10, 860, 10.3390/genes10110860.
- Mizoguchi, T.; Wright, L.; Fujiwara, S.; Cremer, F.; Lee, K.; Onouchi, H.; Mouradov, A.; Fowler, S.; Kamada, H.; Putterill, J.; et al. Distinct Roles of GIGANTEA in Promoting Flowering and Regulating Circadian Rhythms in Arabidopsis. Plant Cell 2005, 17, 2255–2270.
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 2013, 3, 671–677.
- Woe-Yeon Kim; Sumire Fujiwara; Sung-Suk Suh; Jeongsik Kim; Yumi Kim; Linqu Han; Karine David; Joanna Putterill; Hong Gil Nam; David E. Somers; et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356-360, 10.1038/nature06132.
- Jeongsik Kim; Ruishuang Geng; Richard A. Gallenstein; David E. Somers; The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA.. Development 2013, 140, 4060-4069, 10.1242/dev.096651.
- Jae-Woong Yu; Vicente Rubio; Na-Yeoun Lee; Sulan Bai; Sun-Young Lee; Sang-Sook Kim; Lijing Liu; Yiyue Zhang; María Luisa Irigoyen; James A. Sullivan; et al.Yu ZhangIlha LeeQi XieNam-Chon PaekXing Wang Deng COP1 and ELF3 Control Circadian Function and Photoperiodic Flowering by Regulating GI Stability. Molecular Cell 2008, 32, 617-630, 10.1016/j.molcel.2008.09.026.
- Alexandra Pokhilko; Aurora Piñas Fernández; Kieron D Edwards; Megan M Southern; Karen J Halliday; Andrew J Millar; The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Molecular Systems Biology 2012, 8, 574-574, 10.1038/msb.2012.6.
- Sally Adams; Ian Manfield; Peter Stockley; Isabelle A. Carré; Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLOS ONE 2015, 10, e0143943, 10.1371/journal.pone.0143943.
- Paloma Más; Woe-Yeon Kim; David E. Somers; Steve A. Kay; Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567-570, 10.1038/nature02163.
- Mark R. Doyle; Seth J. Davis; Ruth M. Bastow; Harriet G. McWatters; László Kozma-Bognár; Ferenc Nagy; Andrew J Millar; Richard M. Amasino; The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74-77, 10.1038/nature00954.
- Claudio Brandoli; César Petri; Marcos Egea-Cortines; Julia Weiss; The clock gene Gigantea 1 from Petunia hybrida coordinates vegetative growth and inflorescence architecture. Scientific Reports 2020, 10, 1-17, 10.1038/s41598-019-57145-9.
- Joon-Yung Cha; Jeongsik Kim; Tae-Sung Kim; Qingning Zeng; Lei Wang; Sang Yeol Lee; Woe-Yeon Kim; David E. Somers; GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nature Communications 2017, 8, 1-12, 10.1038/s41467-016-0014-9.
- David E Somers; Thomas F Schultz; Maureen Milnamow; Steve A Kay; ZEITLUPE Encodes a Novel Clock-Associated PAS Protein from Arabidopsis. Cell 2000, 101, 319-329, 10.1016/s0092-8674(00)80841-7.
- Marta I. Terry; Fernando Pérez-Sanz; María Victoria Díaz-Galián; Felipe Pérez De Los Cobos; Pedro Navarro; Marcos Egea-Cortines; Julia Weiss; The Petunia CHANEL Gene is a ZEITLUPE Ortholog Coordinating Growth and Scent Profiles.. Cells 2019, 8, 343, 10.3390/cells8040343.
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120.
- Imaizumi, T.; Kay, S.A. Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 2006, 11, 550–558.
- Federico Valverde; Aidyn Mouradov; Wim Soppe; Dean Ravenscroft; Alon Samach; George Coupland; Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science 2004, 303, 1003-1006, 10.1126/science.1091761.
- Mariko Sawa; Dmitri A. Nusinow; Steve A. Kay; Takato Imaizumi; FKF1 and GIGANTEA Complex Formation Is Required for Day-Length Measurement in Arabidopsis. Science 2007, 318, 261-265, 10.1126/science.1146994.
- Takato Imaizumi; Hien G. Tran; Trevor E. Swartz; Winslow R. Briggs; Steve A. Kay; FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 2003, 426, 302-306, 10.1038/nature02090.
- Tong-Seung Tseng; Patrice A. Salomé; C. Robertson McClung; Neil Olszewski; SPINDLY and GIGANTEA Interact and Act in Arabidopsis thaliana Pathways Involved in Light Responses, Flowering, and Rhythms in Cotyledon Movements. The Plant Cell 2004, 16, 1550-1563, 10.1105/tpc.019224.
- Stephen M. Swain; Tong-Seung Tseng; Neil E. Olszewski; Altered Expression of SPINDLY Affects Gibberellin Response and Plant Development. PLANT PHYSIOLOGY 2001, 126, 1174-1185, 10.1104/pp.126.3.1174.
- Mariko Sawa; Steve A. Kay; GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proceedings of the National Academy of Sciences 2011, 108, 11698-11703, 10.1073/pnas.1106771108.
- Jae-Hoon Jung; Yeon-Hee Seo; Pil Joon Seo; Jose Luis Reyes; Ju Yun; Nam-Hai Chua; Chung-Mo Park; The GIGANTEA-Regulated MicroRNA172 Mediates Photoperiodic Flowering Independent of CONSTANS in Arabidopsis. The Plant Cell 2007, 19, 2736-2748, 10.1105/tpc.107.054528.
- Young Hun Song; Akane Kubota; Michael S. Kwon; Michael F. Covington; Nayoung Lee; Ella R. Taagen; Dianne Laboy Cintrón; Dae Yeon Hwang; Reiko Akiyama; Sarah K. Hodge; et al.He HuangNhu H. NguyenDmitri A. NusinowAndrew J MillarKentaro K. ShimizuTakato Imaizumi Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nature Plants 2018, 4, 824-835, 10.1038/s41477-018-0253-3.
- Zhao, X.Y.; Liu, M.S.; Li, J.R.; Guan, C.M.; Zhang, X.S. The wheat TaGI1, involved in photoperiodic flowering, encodesan Arabidopsis GI ortholog. Plant Mol. Biol. 2005, 58, 53–64.
- Hecht, V.; Knowles, C.L.; Vander Schoor, J.K.; Liew, L.C.; Jones, S.E.; Lambert, M.J.M.; Weller, J.L. Pea LATE BLOOMER1 Is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 2007, 144, 648–661.
- Wuriyanghan, H.; Zhang, B.; Cao, W.-H.; Ma, B.; Lei, G.; Liu, Y.-F.; Wei, W.; Wu, H.-J.; Chen, L.-J.; Chen, H.-W.; et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 2009, 21, 1473–1494.
- Bendix, C.; Mendoza, J.M.; Stanley, D.N.; Meeley, R.; Harmon, F.G. The circadian clock-associated gene gigantea1 affects maize developmental transitions: Gigantea1 regulates maize developmental transitions. Plant Cell Environ. 2013, 36, 1379–1390.
- Xie, Q.; Lou, P.; Hermand, V.; Aman, R.; Park, H.J.; Yun, D.-J.; Kim, W.Y.; Salmela, M.J.; Ewers, B.E.; Weinig, C.; et al. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834.
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Lagercrantz, U. Conserved function of core clock proteins in the Gymnosperm Norway spruce (Picea abies L. Karst). PLoS ONE 2013, 8, e60110.
- Tang, W.; Yan, H.; Su, Z.; Park, S.-C.; Liu, Y.; Zhang, Y.; Wang, X.; Kou, M.; Ma, D.; Kwak, S.-S.; et al. Cloning and characterization of a novel GIGANTEA gene in sweet potato. Plant Physiol. Biochem. 2017, 116, 27–35.
- Satoshi Watanabe; Zhengjun Xia; Rumiko Hideshima; Yasutaka Tsubokura; Shusei Sato; Naoki Yamanaka; Ryoji Takahashi; Toyoaki Anai; Satoshi Tabata; Keisuke Kitamura; et al.Kyuya Harada A Map-Based Cloning Strategy Employing a Residual Heterozygous Line Reveals that theGIGANTEAGene Is Involved in Soybean Maturity and Flowering. Genetics 2011, 188, 395-407, 10.1534/genetics.110.125062.
- Yan Wang; Yongzhe Gu; Huihui Gao; Lijuan Qiu; Ruzhen Chang; Shouyi Chen; Chang Ruzhen; Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evolutionary Biology 2016, 16, 1-13, 10.1186/s12862-016-0653-9.
- Tao Wang; Ming-Yang Sun; Xue-Song Wang; Wen-Bin Li; Yong-Guang Li; Over-Expression of GmGIa-Regulated Soybean miR172a Confers Early Flowering in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences 2016, 17, 645, 10.3390/ijms17050645.
- Shakhira Zakhrabekova; Simon P. Gough; Ilka Braumann; André H. Müller; Joakim Lundqvist; Katharina Ahmann; Christoph Dockter; Izabela Matyszczak; Marzena Kurowska; Arnis Druka; et al.Robbie WaughAndreas GranerNils SteinBurkhard SteuernagelUdda LundqvistMats Hansson Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proceedings of the National Academy of Sciences 2012, 109, 4326-4331, 10.1073/pnas.1113009109.
- Hayama, R.; Izawa, T.; Shimamoto, K. Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 2002, 43, 494–504.
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794.
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722.
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, A rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105.
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483.
- Qingbo Ke; Ho Soo Kim; Zhi Wang; Chang Yoon Ji; Jae Cheol Jeong; Haeng-Soon Lee; Young-Im Choi; Bingcheng Xu; Xiping Deng; Dae-Jin Yun; et al.Sang-Soo Kwak Down-regulation ofGIGANTEA-likegenes increases plant growth and salt stress tolerance in poplar. Plant Biotechnology Journal 2016, 15, 331-343, 10.1111/pbi.12628.
- Sandra Rychel; Michał Książkiewicz; Magdalena Tomaszewska; Wojciech Bielski; Bogdan Wolko; FLOWERING LOCUS T, GIGANTEA, SEPALLATA, and FRIGIDA homologs are candidate genes involved in white lupin (Lupinus albus L.) early flowering. Molecular Breeding 2019, 39, 43, 10.1007/s11032-019-0952-0.
- Akane Kubota; Shogo Kita; Kimitsune Ishizaki; Ryuichi Nishihama; Katsuyuki T. Yamato; Takayuki Kohchi; Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nature Communications 2014, 5, 3668, 10.1038/ncomms4668.
- Casal, J.J.; Luccioni, L.G.; Oliverio, K.A.; Boccalandro, H.E. Light, phytochrome signalling and photomorphogenesis in ArabidopsisDedicated to Professor Silvia Braslavsky, to mark her great contribution to photochemistry and photobiology particularly in the field of photothermal methods. Photochem. Photobiol. Sci. 2003, 2, 625.
- Martin-Tryon, E.L.; Kreps, J.A.; Harmer, S.L. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007, 143, 473–486.
- Chentao Lin; Plant blue-light receptors. Trends in Plant Science 2000, 5, 337-342, 10.1016/s1360-1385(00)01687-3.
- Priyanka Emishra; Kishore Chandra Sekhar Panigrahi; GIGANTEA – an emerging story. Frontiers in Plant Science 2015, 6, 8, 10.3389/fpls.2015.00008.
- Karina Andrea Oliverio; María Crepy; Ellen L. Martin-Tryon; Raechel Milich; Stacey L. Harmer; J. Putterill; Marcelo J. Yanovsky; Jorge J. Casal; GIGANTEA Regulates Phytochrome A-Mediated Photomorphogenesis Independently of Its Role in the Circadian Clock. Plant Physiology 2007, 144, 495-502, 10.1104/pp.107.097048.
- Park, Y.-J.; Kim, J.Y.; Lee, J.-H.; Lee, B.-D.; Paek, N.-C.; Park, C.-M. GIGANTEA shapes the photoperiodic rhythms of thermomorphogenic growth in Arabidopsis. Mol. Plant 2020, 13, 459–470.
- Nohales, M.A.; Kay, S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 21893–21899.
- Hwang, C.; Park, J.; Lee, B.; Cheong, H.; Loss of Function in GIGANTEA Gene is Involved in Brassinosteroid Signaling. J. Chosun Nat. Sci. 2011, 4, 113–120.
- Kim, W.-Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.-Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1–13.
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60.
- Matteo Riboni; Alice Robustelli Test; Massimo Galbiati; Chiara Tonelli; Lucio Conti; Environmental stress and flowering time. Plant Signaling & Behavior 2014, 9, e29036, 10.4161/psb.29036.
- Corrales, A.-R.; Nebauer, S.G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.-V.; et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012.
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86
- Joon-Yung Cha; Da-Yeon Lee; Imdad Ali; Song Yi Jeong; BoBae Shin; Hyunjung Ji; Jung Sun Kim; Min-Gab Kim; Woe-Yeon Kim; Arabidopsis GIGANTEA negatively regulates chloroplast biogenesis and resistance to herbicide butafenacil. Plant Cell Reports 2019, 38, 793-801, 10.1007/s00299-019-02409-x.
- Jasmina Kurepa; Jan Smalle; Marc Va; N Montagu; Dirk Inze; Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. The Plant Journal 1998, 14, 759-764, 10.1046/j.1365-313x.1998.00168.x.
- Mohammad Reza Bolouri Moghaddam; Wim Van Den Ende; Sugars, the clock and transition to flowering. Frontiers in Plant Science 2013, 4, null, 10.3389/fpls.2013.00022.
- Neil Dalchau; Seong J. Baek; Helen M. Briggs; Fiona C. Robertson; Antony N. Dodd; Michael J. Gardner; Matthew A. Stancombe; Michael J. Haydon; Guy-Bart Stan; Jorge M. Gonçalves; et al.Alex A. R. Webb The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proceedings of the National Academy of Sciences 2011, 108, 5104-5109, 10.1073/pnas.1015452108.
- Michael J. Haydon; Olga Mielczarek; Alexander Frank; Ángela Román; Alex A.R. Webb; Sucrose and Ethylene Signaling Interact to Modulate the Circadian Clock. Plant Physiology 2017, 175, 947-958, 10.1104/pp.17.00592.
- S. Q. Cao; Y. Q. Song; L. Su; Freezing sensitivity in the gigantea mutant of Arabidopsis is associated with sugar deficiency. Biologia plantarum 2007, 51, 359-362, 10.1007/s10535-007-0073-1.
- Klaus Eimert; Shue-Mei Wang; Wei-Ling Lue; Jychian Chen; Monogenic Recessive Mutations Causing Both Late Floral Initiation and Excess Starch Accumulation in Arabidopsis. The Plant Cell 1995, 7, 1703, 10.2307/3870031.
- Johanna Krahmer; Greg S. Goralogia; Akane Kubota; Argyris Zardilis; Richard S. Johnson; Young Hun Song; Michael J. MacCoss; Thierry Ele Bihan; Karen J. Halliday; Takato Imaizumi; et al.Andrew J. Millar Time‐resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Letters 2018, 593, 319-338, 10.1002/1873-3468.13311.
- Gulzar S. Sanghera; Shabir H. Wani; Wasim Hussain; N. B. Singh; Engineering Cold Stress Tolerance in Crop Plants. Current Genomics 2011, 12, 30-43, 10.2174/138920211794520178.
- C. Guy; Cold Acclimation And Freezing Stress Tolerance: Role Of Protein Metabolism. Annual Review of Plant Biology 1990, 41, 187-223, 10.1146/annurev.arplant.41.1.187.
- J. Weiss; Marcos Egea-Cortines; Transcriptomic analysis of cold response in tomato fruits identifies dehydrin as a marker of cold stress. Journal of Applied Genetics 2009, 50, 311-319, 10.1007/bf03195689.
- Guy, C.L.; Niemi, K.J.; Brambl, R. Altered gene expression during cold acclimation of spinach. Proc. Natl. Acad. Sci. USA 1985, 82, 3673–3677.
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599.
- Matsuo Uemura; Gareth Warren; Peter L. Steponkus; Freezing Sensitivity in the sfr4 Mutant of Arabidopsis Is Due to Low Sugar Content and Is Manifested by Loss of Osmotic Responsiveness. Plant Physiology 2003, 131, 1800-1807, 10.1104/pp.102.013227.
- M. Miquel; D. James; H. Dooner; J. Browse; Arabidopsis requires polyunsaturated lipids for low-temperature survival.. Proceedings of the National Academy of Sciences 1993, 90, 6208-6212, 10.1073/pnas.90.13.6208.
- Z. Ristic; E. N. Ashworth; Changes in leaf ultrastructure and carbohydrates inArabidopsis thaliana L. (Heyn) cv. Columbia during rapid cold acclimation. Protoplasma 1993, 172, 111-123, 10.1007/bf01379368.
- Perla Gómez; M Ángeles Ferrer; J Pablo Fernández-Trujillo; Antonio Calderón; Francisco Artés; Marcos Egea-Cortines; Julia Weiss; Structural changes, chemical composition and antioxidant activity of cherry tomato fruits (cv. Micro-Tom) stored under optimal and chilling conditions. Journal of the Science of Food and Agriculture 2009, 89, 1543-1551, 10.1002/jsfa.3622.
- Immink, R.G.; Hannapel, D.J.; Ferrario, S.; Busscher, M.; Franken, J.; Lookeren Campagne, M.M.; Angenent, G.C. A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 1999, 126, 5117–5126.
- Souer, E.; Rebocho, A.B.; Bliek, M.; Kusters, E.; de Bruin, R.A.M.; Koes, R. Patterning of Inflorescences and Flowers by the F-Box Protein DOUBLE TOP and the LEAFY Homolog ABERRANT LEAF AND FLOWER of Petunia. Plant Cell 2008, 20, 2033–2048
- Knudsen, J.T.; Tollsten, L.; Bergström, L.G. Floral scents—A checklist of volatile compounds isolated by head-space techniques. Phytochemistry 1993, 33, 253–280.
- Pichersky, E.; Gershenzon, J. The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 2002, 5, 237–243.
- Natalia Dudareva; Antje Klempien; Joëlle K. Muhlemann; Ian Kaplan; Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytologist 2013, 198, 16-32, 10.1111/nph.12145.
- Alain Tissier; John A Morgan; Natalia Dudareva; Plant Volatiles: Going ‘In’ but not ‘Out’ of Trichome Cavities. Trends in Plant Science 2017, 22, 930-938, 10.1016/j.tplants.2017.09.001.
- Efraim Lewinsohn; Nativ Dudai; Yaakov Tadmor; Irena Katzir; Uzi Ravid; Eli Putievsky; Daniel M. Joel; Histochemical Localization of Citral Accumulation in Lemongrass Leaves (Cymbopogon citratus(DC.) Stapf., Poaceae). Annals of Botany 1998, 81, 35-39, 10.1006/anbo.1997.0525.
- Vincent R. Franceschi; Paal Krokene; Erik Christiansen; Trygve Krekling; Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytologist 2005, 167, 353-376, 10.1111/j.1469-8137.2005.01436.x.
- Colquhoun, T.A.; Verdonk, J.C.; Schimmel, B.C.J.; Tieman, D.M.; Underwood, B.A.; Clark, D.G. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry 2010, 71, 158–167.
- Cna’Ani, A.; Mühlemann, J.K.; Ravid, J.; Masci, T.; Klempien, A.; Nguyen, T.T.H.; Dudareva, N.; Pichersky, E.; Vainstein, A. Petunia × hybrida floral scent production is negatively affected by high-temperature growth conditions: Ambient temperature and floral scent. Plant Cell Environ. 2015, 38, 1333–1346.
- Myles P. Fenske; Kristen D. Hewett Hazelton; Andrew K. Hempton; Jae Sung Shim; Breanne M. Yamamoto; Jeffrey A. Riffell; Takato Imaizumi; Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia. Proceedings of the National Academy of Sciences 2015, 112, 9775-9780, 10.1073/pnas.1422875112.


