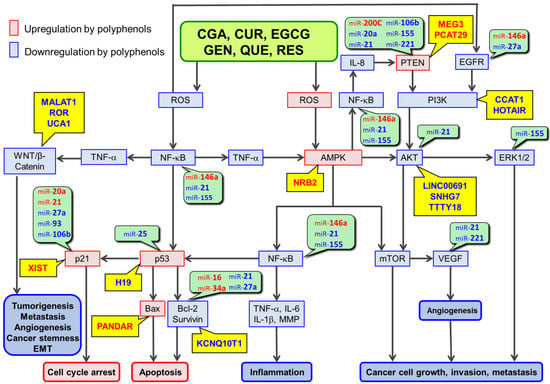
The anticancer effects of daily consumption of polyphenols. These dietary polyphenols include chlorogenic acid, curcumin, epigallocatechin-3-O-gallate, genistein, quercetin, and resveratrol. These polyphenols have similar chemical and biological properties in that they can act as antioxidants and exert the anticancer effects via cell signaling pathways involving their reactive oxygen species (ROS)-scavenging activity. These polyphenols may also act as pro-oxidants under certain conditions, especially at high concentrations. Epigenetic modifications, including dysregulation of noncoding RNAs (ncRNAs) such as microRNAs, long noncoding RNAs, and circular RNAs are now known to be involved in the anticancer effects of polyphenols. These polyphenols can modulate the expression/activity of the component molecules in ROS-scavenger-triggered anticancer pathways (RSTAPs) by increasing the expression of tumor-suppressive ncRNAs and decreasing the expression of oncogenic ncRNAs in general. Multiple ncRNAs are similarly modulated by multiple polyphenols. Many of the targets of ncRNAs affected by these polyphenols are components of RSTAPs. Therefore, ncRNA modulation may enhance the anticancer effects of polyphenols via RSTAPs in an additive or synergistic manner, although other mechanisms may be operating as well.
- anticancer
- ROS
- polyphenols
1. Introduction
| miRs | miR-16 | miR-22 | miR-34a | miR-141 | miR-145 | miR-146a | miR-200c |
|---|---|---|---|---|---|---|---|
| CUR |
| CGA | CUR | EGCG | GEN | QUE | RES | ||
|---|---|---|---|---|---|---|---|
| EGCG | GEN | QUE | RES | ||||
| Polyphenols | |||||||
| miR-7 SET8↓, Bcl-2↓, p53↑ [80]; Skp2↓, p57↑, p21↑ [81] miR-9 AKT↓, FOXO1↓ [82]; GSK-3β↑, β-catenin↑, Cyclin D1↓ [83] miR-15a Bcl-2↓ [9]; WT1↓ [84] miR-16-1 WT1↓ [84] miR-28-5p BECN1↓ [85] miR-29a DNMT1↓, 3A↓, 3B↓ [86] miR-30c-5p MTA1↓ [87] miR-33b HMGA2↓ [88]; XIAP↓ [89] miR-98 LIN28A↓, MMP2↓, MMP9↓ [90] miR-99a JAK1↓, STAT1↓, STAT3↓ [91] | CUR Yang et al. [9] EGCG Tsang et al.[10] QUE STsang et al. [10] QUE Sonoki et al. [11]; Zhaonoki et al. [11];[12 | : Hagiwara et al. | [ | 13 | ] | EMT↓ via vimentin, ZEB1↑, E-cadherin↓ RES: Dermani et al. [45] |
| miRs | miR-20a | miR-21 | miR-25 | miR-27a | miR-93 | miR-106b | miR-155 | miR-221 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-101 EZH2↓, EpCAM↓ [92]; Notch1↓ [93]; EZH2↓ [94] miR-124 Midkine↓ [95] | ] Zhao et al. [12]RES RESHagiwara et al. [13]; HagAzimiwara et al. [13]; Azimi et al. [14][14] |
|||||||||||||
| Polyphenols | CGA Huang et al. [46] CUR Gandhy et al. [47] EGCG Mirzaaghaei et al. [ |
miR-125a ERRα↓ [] |
||||||||||||
| miR-17 p21↑, G0/G1 arrest↑ [46] | 48] 96RES Dhar et al. [49]; Dhar et al. [50]] miR-138 Smad4↓, NF-kB↓, Cyclin D3↓ [97] miR-143 NF-kB↓ [98]; PGK1↓ [99]; Autophagy via ATG2B↓ [100] miR-181b CXCL1↓ [101] miR-185 DNMT1↓, 3A↓, 3B↓ [86] miR-192-5p XIAP↓ [102]; PI3K↓, AKT↓ [103]; Wnt/β-catenin↓ [104] miR-196b ** BCR-ABL↓ [55] miR-206 mTOR↓, AKT↓ [105] miR-215 XIAP↓ [102] miR-340 XIAP↓ [106] miR-384 circ-PRKCA↓ [107] miR-491 PEG10↓ [108] miR-593 MDR1↓ [109 | CUR Sun et al. [15 |
miR-19a,b PTEN↑ [131] miR-125a-5p TP53↑ [132] miR-130a Nkd2↑ [133] miR-7641 p16↑ [134]]; Sreenivasan et al. [16]; Sibbesen et al. [17] EGCG Li et al. [18] QUE Zhang et al. [19] |
CGA Wang et al. [51] CUR Mudduluru et al. [52]; Subramaniam et al. [53]; Zhang et al. [54]; Taverna et al. [55]; Yallapu et al. [56] EGCG Fix et al. [57] **; Siddiqui et al. [58] GEN Zaman et al. [59] RES Tili et al. [60]; Sheth et al. [61]; Liu et al. [62]; Li et al. [63]; Zhou et al. [64]miR-15b STIM2↓, Orai1↓ [110] miR-29b KDM2A↓ [111] miR-485-5p RXRα↓ [112] let-7b HMGA2↓ [113] |
miR-98-5p CTR1↑ [135]CUR Guo et al. [20]; Sun et al. [21]; Toden et al. [22]; Sun et al. [15] EGCG Chakrabarti et al. [23]; Li et al. [18]; Chakrabarti et al. [24]; Toden et al. [25]; Mostafa et al. [26] GEN Hsieh et al. [27]; Xia et al. [ |
CUR28]; Chiyomaru et al. [29] RES Hagiwara et al. [13]; Otsuka et al. [30]; Kumazaki et al. [31]; Yao et al. [32] |
Sun et al. [15] EGCG Fix et al. [57] **; Gordon et al. [34]; Zan et al. [65] RES Tili et al. [60]miR-574-3p RAC1↓, EGFR↓, EP300↓ [114] miR-1469 Mcl1↓ [115] let-7d THBS1↓ [116] |
miR-23b-3p PTEN↑ [136] miR-151a-5p CASZ1↑, IL1RAPL1↑, SOX17↑, N4BP1↑, ARHGDIA↑ [137] miR-155 PTEN↑ [73] miR-221 miR-222 ARHI↑ [78] miR-223 Fbw7↑ [138] miR-223 E-cadherin↑ [139] miR-873-5p FOXM1↓ [140] miR-1260b sFRP1↑, Smad4↑, Dkk2↑ [141][142][141,142CUR Toden et al. [33] EGCG Gordon et al. [34] GEN Chiyomaru et al. [35] RES Hagiwara et al. [13] |
]CUR Toden et al. [22]; Noratto et al. [66] EGCG Fix et al. [57] ** GEN Xia et al. [miR-1-3p TAGLN2↓ [11767]; Xu et al. [68]; Sun et al. [69]] miR-16-5p WEE1↓ [118] miR-22 Wnt1↓ [19] miR-34a-5p SNHG7↓ [119] miR-142-3p HSP70 ↓ [120] miR-197 IGFBP5↓ [121] miR-200b-3p Notch1↓ [122] miR-217 KRAS↓ [123] miR-503-5p Cyclin D1↓ [124] miR-1254 CD36↓ [125] miR-1275 IGF2BP1↓, IGF2BP3↓ [126] let7-a KRAS↓ [127] let-7c Numbl/Notch1↑ [ |
miR-30d-5p Notch↓ Wnt↓ [143128]CUR Mirgani et al. [36]; Liu et al. [37] EGCG Toden et al. [25] GEN Wei et al. [38] QUE Zhou et al. [39] RES Sachdeva et al. [40] |
CGA Huang et al. [46] EGCG Chakrabarti et al. [23]; Chakrabarti et al. [24] RES Singh et al. [70] |
miR-424-3p Galectin-3↓ [129]CUR Wu et al. [41] GEN Li et al. [42] QUE Tao et al. [43] |
CGA Huang et al. [46] EGCG Chakrabarti et al. [23] RES Dhar et al. [50]; Dhar et al. [49]CUR Toden et al. [33]; Soubani et al. [44] EGCG Toden et al. [25] RES Hagiwara et al. [13]; Dermani et al. [ |
CGA Zeng et al. [7145] |
| ] | CUR | Ma et al. | [ | 72 | ] GEN de la Parra et al. [73] QUE Boesch-Saadatmandi et al. [74] RES Tili et al. [75] |
CUR Zhang et al. [76]; Allegri et al. [77] EGCG Allegri et al. [77] GEN Chen et al. [78] |
Targets * | Bcl-2↓ CUR: Yang et al. [9]; EGCG: Tsang et al. [10] HOXA10↓ QUE: Zhao et al. [12] |
||||||
| Targets * | p21↑ CGA: Huang et al. [46] PTEN↑ RES: Dhar et al. [49] | SP1↓, ESR1↓ CUR: Sun et al. [15] Erbb3↓ CUR: Sreenivasan et al. [16] NCoA1↓, HDAC6↓, MYCBP↓, PTEN↓, CUR: Sibbesen et al. [17] Wnt1/β-catenin↓ QUE: Zhang et al. [19] |
Smad7↑ CGA: Wang et al. [51] PDCD4↑ CUR: Mudduluru et al. [52] PTEN↑ CUR: Zhang et al. [54] PTEN↑ CUR: Taverna et al. [55] p21↑, p38 MAPK↑, Cyclin E2↓ GEN: Zaman et al. [59] PDCD4↑ RES: Sheth et al. [61] Bcl-2↓ RES: Liu et al. [62] NF-κB↓ RES: Liu et al. [63] AKT↓, Bcl-2↓ RES: Zhou et al. [64]Bcl-2↓, Bmi-1↓ CUR: Guo et al. [20] Bcl-2↓, CDK4↓, Cyclin D1↓ CUR: Sun et al. [21] Cyclin D↓, c-Myc↓, CDK6↓, Bcl-2↓ CUR: Toden et al. [22] miR-92↓, miR-93↓, miR-106b↓, miR-7-1↑, miR-34a↑, miR-99a↑ EGCG: Chakrabarti et al. [23] EMT↓, RTCB↓, ROS↑ GEN: Hsieh et al. [27] HOTAIR↓ GEN: Chiyomaru et al. [29] Notch-1↓ GEN: Xia et al. [28] Sirt1↓ via E2F3 RES: Kumazaki et al. [31] HNRNPA1↓ |
p53↑ EGCGRES: Otsuka et al. [30] Bcl-2↓ RES: Yao et al. [32] |
: Gordon et al. [34] PARP1↑, Caspase 3↑, Caspase 9↑ EGCG: Zan et al. [65]EMT↓ CUR: Toden et al. [33] HOTAIR↓ GEN: Chiyomaru et al. [35] Cancer stemness↓ |
Cyclin E1↓, c-Myc↓ via FBXW7 CURRES: Hagiwara et al. [13] |
: Toden et al. [22] ZBTB10-Sp↑ CUR: Noratto et al. [66] Sp1↓, Sp3↓ Sp4↓, EGFR↓, hepatocyte growth factor receptor↓, survivin↓, Bcl-2↓, Cyclin D1↓, NFκB↓, ZBTB4↑ CUR: Grandhy et al. [47] Spry2↑ GEN: Xu et al. [68]Oct4↓, SOX-2↓, Oct4B1↓, CUR: Mirgani et al. [36] Oct4↓, CD44↓, CD133↓, Cyclin D1↓, Cdk4↓ CUR: Liu et al. [37] c-Myc↓ EGCG: Toden et al. [25] ABCE1↓ GEN: Wei et al. [38] Caspase 3↑ QUE: Zhou et al. [39] |
p21↑ CGA: Huang et al. [46] Caspase 8↑, tBid↑, Calpain↑, Caspase 3↑ EGCG: Chakrabarti et al. [23]NF-κB↓ CUR: Wu et al. [41] EGFR↓ MTA-2↓, IRAK-1↓, NF-κB↓ GEN: Li et al. [ |
p21↑ CGA42] Caspase 3↑, Bax↑, EGFR↓ QUE: Tao et al. [43] |
: Huang et al. [46] PTEN↑ RES: Dhar et al. [50]; Dhar et al. [49]EMT↓ CUR: Toden et al. [33] PTEN↑, MT1-MMP↓ CUR: Soubani et al. [44] Cancer stemness↓ EGCG: Toden et al. [25] Cancer stemness↓ RES |
Inflammation↓ via NF-κB/NLRP3 CGA: Zeng et al. [71] SOCS1↓, IL-6↓, CUR: Ma et al. [72] PTEN↑, FOXO3a↑ GEN: de la Parra et al. [73] AP-1↓ via miR-663 RES: Tili et al. [75] |
PTEN↑, p27↑, p57↑, PUMA↑ CUR: Sarkar et al. [79] FGF2↓, MMP2↓, VEGF↓, HGF↓, CUR: Zhang et al. [76] miR-21↓, miR-146b↓, miR-221↓, miR-222↓ CUR: Allegri et al. [77] miR-221↓, EGCG: Allegri et al. [77] ARHI↑ GEN:Chen et al. [78] |
| ] | |||||
| miR-196b ** | miR-1290 | IGFBP3↑ | [130] |
