Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 2 by Rita Xu.
Global water scarcity has led to significant dependence on reclaimed or recycled water for potable uses. Effluents arising from human and animal gut microbiomes highly influence water quality. Wastewater pollution is, therefore, frequently monitored using bacterial indicators (BI).
- wastewater
- indicators
- management
- pepper mild mottle virus
- enteric viruses
- Public health
1. Introduction
The wastewater virome is a distinct subset of the microbiome owing to frequent infection of humans and domestic/companion animals [1][2][3][4]. For instance, a typical healthy human is estimated to harbor more than 10 virus-mediated chronic infections and occasionally more [5]. In particular, the human gut virome composition is intensively studied because of the continuous introduction of newly pathogenic agents and altered pathogenesis patterns, along with changes in immune responses owing to selection pressure imposed on the existent virome [6][7]. Gastroenteritis displays the potential pathogenesis of acquired gut virome [3][8]. For instance, rotavirus A, noroviruses and astroviruses are considered as the major causes of acute gastroenteritis worldwide and mainly result in infantile acute diarrhea [9][10][11].
Exposure to wastewater represents a common transmission route of enteric viruses via recreation water, surface water usage, wastewater/greywater-mediated irrigation and toilet flushing [12][13][14][15][16]. Therefore, wastewater reuse guidelines were suggested for safe use of wastewater. Moreover, fecal contamination indicators were proposed to ensure compliance with these guidelines. Coliform members represent the frequently used fecal indicators; however, other indicators have also been proposed involving bacteriophages, enterococci and sulfite-reducing bacteria [17][18][19][20]. These have not, however, met the expected sensitivity of enteric viruses’ detection [21]. On the contrary, microbial source tracking (MST) tools provided higher specificity. Currently, MST professionals adopt a toolbox approach, i.e., implementing numerous MST markers, such as using pepper mild mottle virus (PMMoV) together with cross-assembly phage crAssphage [22][23][24].
2. Viral Contamination of Treated Wastewater
Viruses are obligatory intracellular parasites of both eukaryotic and prokaryotic cells, and considered as the smallest microorganisms capable of replication [25]. Since they are unable to metabolize, they cannot engage in any energy-dependent processes like growth, respiration or reproduction on their own [26]. The main component of viruses is nucleic acid (DNA or RNA), which is shielded by a protein capsid that, in some cases, is surrounded by a lipid envelope [27]. Despite their apparent simplicity, viruses can nevertheless penetrate host cells using a number of physical and chemical mechanisms that are part of both the virus and the cell’s structure. Moreover, viruses can also manipulate cellular processes to produce progeny viruses using various routes of entry [28]. Of particular interest is that enteric viruses are transmitted via the fecal–oral pathway. Enteric viruses are considered as the most persistent fecal microorganisms even during treatment approaches of contaminated raw water owing to their unique characteristics [29]. These viruses have an icosahedral structure, are between 20 and 100 nm in size, and primarily show a negative charge at neutral pH [30]. They are principal causes of viral gastroenteritis, hepatitis and poliomyelitis, displaying their adverse effect on public health. In addition, some enteric viruses, including polyomaviruses, have been linked to cancer [31]. Moreover, enteric viruses are highly efficient at surviving outside the gut for long periods, thus are easily disseminated through water resources [32]. Inappropriate wastewater treatment has caused viral contamination of shellfish, fresh produce and recreational waterways [33]. Many developing nations struggle with this ongoing problem because they lack the resources for effective wastewater treatment [34]. It is not surprising that, in current US frameworks, viruses demand substantially bigger reductions than bacteria or protozoan parasites when wastewater is reused for potable reasons. Target log10 removal value (LRV) attributions are computed using a risk-modelling methodology, assuming the worst-case scenario (very high viral concentrations, as would be the situation during a big epidemic), and a final risk of less than one illness in 10,000 exposures, per year [35]. Currently, states are choosing between one of these three LRV programs that were previously developed [36]. Although Texas’ requirements seem to be less stringent than those of California or the National Water Research Institute (NWRI), it should be emphasized that Texas only counts LRVs from treated effluent toward final product water, excluding wastewater treatment reduction [37]. The wastewater treatment plant must exhibit its ability to eliminate pathogens to the levels required by the state prior to earning an LRV attribution, typically through a pilot demonstration [38]. Validating virus elimination is necessary for LRV attribution in reuse schemes. However, pathogenic virus concentrations in sewage and treated effluent vary, and frequently are not at levels that might effectively verify an 8–12 LRV [39]. It is frequently not practical or safe to spike pathogenic viruses at each phase to confirm overall LRV. Non-pathogenic viruses are frequently used as a process indicator [40].3. Human Health Risk of Virus-Associated Water Pollution
On an annual basis, there are over 4 billion instances of waterborne diarrheal illnesses, which cause 2 million deaths, with under-five year olds the majority [41]. Enteric viral infections account for a sizable fraction of these diseases [42]. The most crucial way for enteric viruses to spread is through direct contact with infected individuals. Enteric viruses are spreading via the fecal–oral pathway as shown in Figure 1 [43]. However, the majority of enteric viruses remain persistent in areas where residential wastewater discharges exist and are frequently linked to waterborne epidemics [43]. Although typical wastewater treatment techniques can be comparatively inefficient at eliminating enteric viruses, wastewater is frequently treated before being released into the environment [44]. Fecal matter pollutes the environment and drinking water sources in poor countries since many locations lack suitable sanitary infrastructure and wastewater treatment facilities [45]. Additionally, significant amounts of untreated wastewater may be released by combined sewer overflows (CSOs) during periods of high rainfall as well as through dry water overflows, such as those caused by snowmelt, tidal infiltration, system failures and obstructions [46][47][48]. Consequently, people who come into direct or indirect contact with contaminated waters are prone to the risk of contracting viral infections as a result of these events, which allow enteric pathogens to contaminate the environment directly [49]. Enteric viruses are extremely contagious in ambient waters and can stick to particles in the water column or accumulate in sediment [50]. They might subsequently be consumed by aquatic organisms, such as bivalve shellfish harvested for human consumption [51]. Additionally, wastewater is regularly used for irrigation in areas with a shortage of freshwater; as a result, enteric viruses may directly contaminate fruit and salad vegetables, and result in foodborne outbreaks [47]. The typical duration of gastroenteritis caused by enteric viruses is 2–5 days [52]. In certain circumstances, the infection goes asymptomatic or causes symptoms in the skin, neurological system or respiratory system [53]. The Picornaviridae, Caliciviridae, Reoviridae, and Adenoviridae families make up the majority of those responsible for gastroenteritis (Table 1). For instance, noroviruses (family Caliciviridae) account for a sizable portion of gastroenteritis infections worldwide, causing 685 million cases and roughly 200,000 fatalities [54], with a total direct cost to the healthcare system of USD 4.2 billion and associated societal costs of USD 60.3 billion annually [54]. The main etiological agents of gastroenteritis in newborns and young children are rotaviruses (family Reoviridae) and group F mastadenoviruses (AdVs; family Adenoviridae) [55]. The three most frequent viral pathogens linked to waterborne and water-associated foodborne outbreaks are noroviruses, hepatitis A virus (family Picornaviridae) and AdVs [56]. Infection can cause significant illness, such as acute hepatitis [57].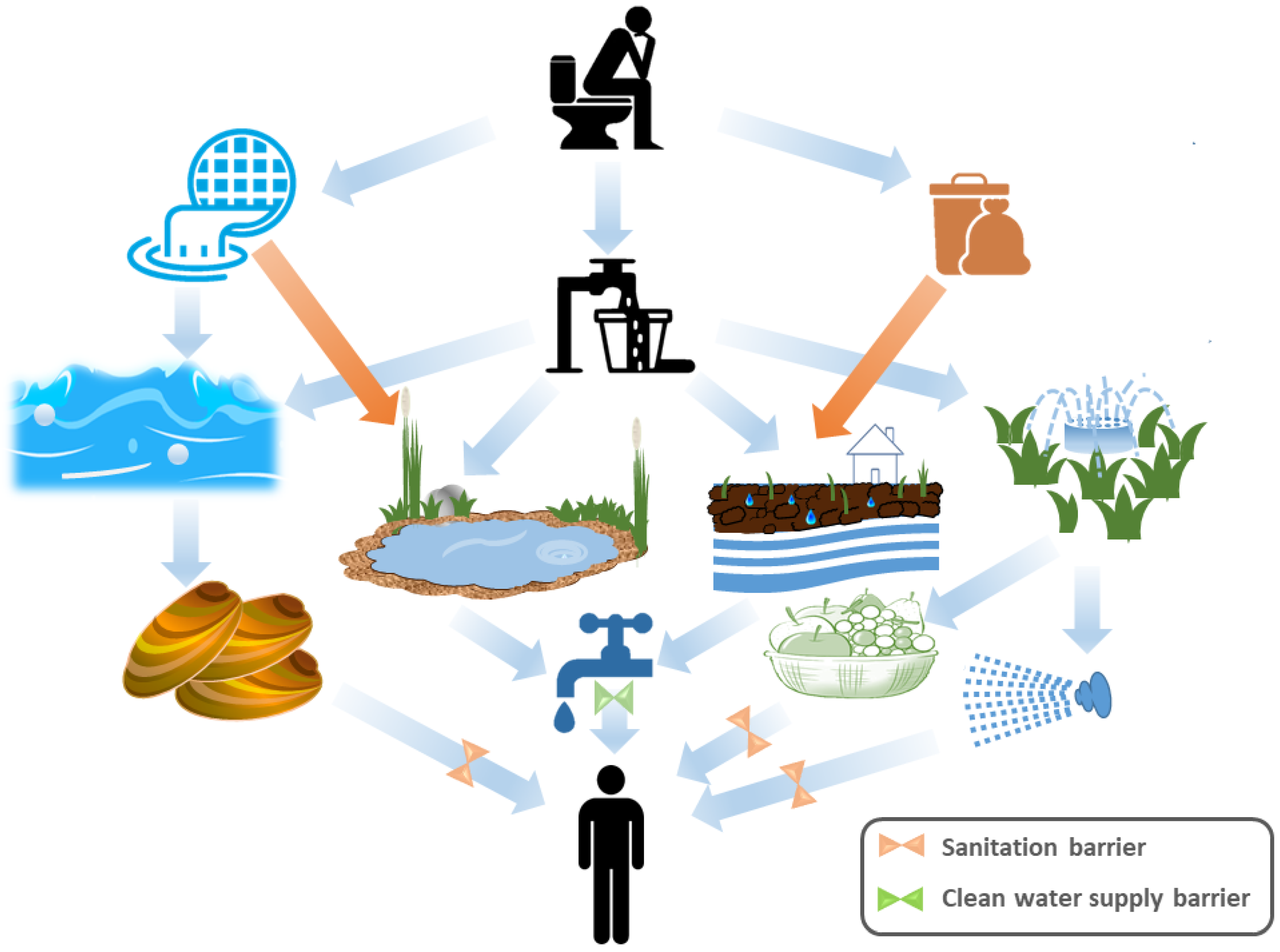
Figure 1. Diagrammatic representation of the fecal–oral route for transmission of enteric viruses. Human excreta go through land runoff and sewage that contaminate oceans, rivers, lakes and ground water. Moreover, sewage can contaminate irrigation water. The contaminated oceans, rivers and lakes influence filter feeders (shellfish) and recreation water, whereas the direct water supply would be affected by the improperly decontaminated ground water and rivers. Crops and irrigation-based aerosols are also contaminated by inadequately treated irrigation water. On the other hand, the human excreta give rise to solid wastes that affect the groundwater, leading to unclean water supply. Absence of a sanitation barrier and a properly clean water supply barrier lead to enteric virus infection of a new human host.
Table 1. Human pathogenic viruses detected in the aquatic environment.
| Virus | Size of Viral Particle | Zoonotic Transmission | Aquatic Environment | References |
|---|---|---|---|---|
| Mastadenovirus A–F | 70–90 nm | No | Wastewater | [58][59] |
| Torque teno virus | 30 nm | Yes | River | [58][59][60] |
| Astrovirus | 28–30 nm | Potentially | Sewage water | [58][61][62] |
| Norovirus GI, GII | 35–40 nm | No | River | [58][63] |
| Sapovirus GI, GII | No | Wastewater and river | [58][64] | |
| Human-associated circovirus | 15–25 nm | No | Sewage | [65][66] |
| Hepatitis E virus type 1–4 | 27–34 nm | Yes | Tap and bottled water | [67][68] |
| Assorted papillomaviruses | 55 nm | No | Wastewater | [69][70] |
| Human bocavirus type 1–4 | 22 nm | No | Recycled water and sewage | [58][71] |
| Aichivirus A–B | 30–32 nm | No | Sewage and surface water | [72] |
| Cosavirus A | No | River and waste water | [65][73] | |
| Coxsackievirus B | No | Sewage water | [73][74] | |
| Enterovirus A–D | No | Groundwater | [73][75] | |
| Poliovirus type 1–3 | No | Wastewater | [73][76] | |
| Hepatitis A virus | 40–45 nm | No | Wastewater | [73][77] |
| BK polyomavirus | No | River and sewage water | [78] | |
| JC polyomavirus | No | Wastewater | [79] | |
| Rotavirus A | 60–80 nm | Potentially | Drinking water | [58][80] |
4. Management Strategies for Wastewater Pollution
4.1. Traditional Fecal Bacterial Indicators
The microbiological safety of irrigation water is monitored using indicator organisms [94]. E. coli is classified as specifically having fecal origin and is a member of the coliform subgroup known as the fecal coliforms [95]. The primary indicator of fecal contamination of water is frequently E. coli [96]. There are several problems with E. coli as a fecal indicator. To begin with, the presence of viral infections is not correlated with E. coli which is not host-specific. Moreover, E. coli also decays in the environment more quickly compared to other foodborne bacteria [97]. In contrast, the standard fecal indicator should show environmental survival and movement across the matrix that are equal to or greater than those of the pathogen, exist at higher concentrations than the pathogen, and provide source specificity [98]. Also, the indicator organism assay method should be accurate, specific, quick, quantitative, sensitive, widely applicable and indicative of infectivity [99]. Levels of fecal contamination in water have conventionally been assessed using fecal indicator bacteria (FIB; including coliform bacteria, Enterococcus, E. coli and Streptococcus spp.) [100]. Bacterial pathogens, like fecal coliforms, can survive for up to 15 days on the surface of food and up to 30 days in water and sewage [101]. However, bacteria have been demonstrated to be substantially less persistent in the environment and significantly less resistant to wastewater treatment than enteric viruses [25]. Consequently, FIB are subpar predictors of the risk of viral infection, which implies that current water-quality monitoring programs based only on FIB are insufficient [102].4.2. Viral Indicators
Human enteric viruses come in about 100 different varieties and the number is growing due to newly discovered and emerging strains [103]. Surrogates and indicators are frequently employed to study the fate and transport of pathogenic strains in the environment owing to the high diversity of viral pathogens [104]. An indicator may be useful for evaluating pathogen abundance, persistence, adsorption and transit in the aquatic environment, as well as for making a general assessment of the effectiveness of wastewater and drinking water treatment [38]. Therefore, a good viral indicator should ideally have comparable inactivation and retention of the target pathogens and should be present year-round in wastewater and habitats impacted by wastewater [105]. This would allow for ongoing monitoring and provide information on the degree of pollution and the probability that pathogens are present [106]. Table 2 lists some enteric viruses that are connected to wastewater and may be utilized as indicators, but not all of these viruses meet the criteria. High concentrations of influenza, corona-, circo- and papillomaviruses have been found in wastewater but not in contaminated areas, which may be because of how quickly they degrade in water [107].Table 2. Survival of enteric viruses in various water environments.
| Organism | Habitat | Temperature | Duration (Days) | Log Reduction | Reference |
|---|---|---|---|---|---|
| Adenovirus | Groundwater | 4 | 132 | 1.00 | [108] |
| 20 | 36 | 1.00 | |||
| Adenovirus 40 | Seawater | 15 | 28 | 1.40 | [109] |
| 15 | 85 | 2.00 | |||
| Drinking water | 4 | 60 | 0.49 | ||
| 4 | 92 | 2.00 | |||
| Adenovirus 41 | Seawater | 15 | 28 | 1.60 | [109] |
| 15 | 77 | 2.00 | |||
| Drinking water | 4 | 60 | 1.00 | ||
| 4 | 304 | 2.00 | |||
| Rotavirus | Fresh water | 20 | 10 | 2.00 | [110] |
| 4 | 32 | 2.00 | |||
| Seawater | 37 | 7 | 5.00 | [111] | |
| Soil | 37 | 7 | 1.70 | [112] | |
| Drinking water | 20 | 64 | 2.00 | [113] | |
| Norovirus | Groundwater | 25 | 1266 | 1.79 | [114] |
| Mineral water | 25 | 80 | 1.30 | [115] | |
| 4 | 80 | 0.89 | |||
| Tap water | 25 | 80 | 0.80 | ||
| 4 | 80 | 3.00 | |||
| Hepatitis A virus | Seawater | 20 | 28 | 4.00 | [116] |
| Artificial seawater | 25 | 11 | 1.00 | [117] | |
| 24 | 19 | 1.00 | |||
| Drinking water | 4 | 60 | 1.60 | [109] | |
| 4 | 56 | 2.00 | |||
| Bottled water | 21 | 21 | 1.99 | [118] | |
| Astrovirus | Tap water | 20 | 30 | 2.00 | [119] |
| 4 | 60 | 2.00 |
References
- Karst, S.M. Viral Safeguard: The Enteric Virome Protects against Gut Inflammation. Immunity 2016, 44, 715–718.
- Majumder, A.; Gupta, A.K.; Ghosal, P.S.; Varma, M. A Review on Hospital Wastewater Treatment: A Special Emphasis on Occurrence and Removal of Pharmaceutically Active Compounds, Resistant Microorganisms, and SARS-CoV-2. J. Environ. Chem. Eng. 2021, 9, 104812.
- Virgin, H.W. The Virome in Mammalian Physiology and Disease. Cell 2014, 157, 142–150.
- Minot, S.; Grunberg, S.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Hypervariable Loci in the Human Gut Virome. Proc. Natl. Acad. Sci. USA 2012, 109, 3962–3966.
- Virgin, H.W.; Wherry, E.J.; Ahmed, R. Redefining Chronic Viral Infection. Cell 2009, 138, 30–50.
- Minot, S.; Bryson, A.; Chehoud, C.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Rapid Evolution of the Human Gut Virome. Proc. Natl. Acad. Sci. USA 2013, 110, 12450–12455.
- Neil, J.A.; Cadwell, K. The Intestinal Virome and Immunity. J. Immunol. 2018, 201, 1615–1624.
- Cadwell, K. The Virome in Host Health and Disease. Immunity 2015, 42, 805–813.
- Espul, C.; Martínez, N.; Noel, J.S.; Cuello, H.; Abrile, C.; Grucci, S.; Glass, R.; Berke, T.; Matson, D.O. Prevalence and Characterization of Astroviruses in Argentinean Children with Acute Gastroenteritis. J. Med. Virol. 2004, 72, 75–82.
- Lartey, B.L.; Quaye, O.; Damanka, S.A.; Agbemabiese, C.A.; Armachie, J.; Dennis, F.E.; Enweronu-Laryea, C.; Armah, G.E. Understanding Pediatric Norovirus Epidemiology: A Decade of Study among Ghanaian Children. Viruses 2020, 12, 1321.
- Phan, T.; Ide, T.; Komoto, S.; Khamrin, P.; Pham, N.T.K.; Okitsu, S.; Taniguchi, K.; Nishimura, S.; Maneekarn, N.; Hayakawa, S. Genomic Analysis of Group A Rotavirus G12P Including a New Japanese Strain Revealed Evidence for Intergenotypic Recombination in VP7 and VP4 Genes. Infect. Genet. Evol. 2021, 87, 104656.
- Gorgich, M.; Mata, T.M.; Martins, A.; Caetano, N.S.; Formigo, N. Application of Domestic Greywater for Irrigating Agricultural Products: A Brief Study. Energy Rep. 2020, 6, 811–817.
- Angelakis, A.N.; Asano, T.; Bahri, A.; Jimenez, B.E.; Tchobanoglous, G. Water Reuse: From Ancient to Modern Times and the Future. Front. Environ. Sci. 2018, 6, 26.
- Libutti, A.; Gatta, G.; Gagliardi, A.; Vergine, P.; Pollice, A.; Beneduce, L.; Disciglio, G.; Tarantino, E. Agro-Industrial Wastewater Reuse for Irrigation of a Vegetable Crop Succession under Mediterranean Conditions. Agric. Water Manag. 2018, 196, 1–14.
- Fountoulakis, M.S.; Markakis, N.; Petousi, I.; Manios, T. Single House On-Site Grey Water Treatment Using a Submerged Membrane Bioreactor for Toilet Flushing. Sci. Total Environ. 2016, 551, 706–711.
- Becerra-Castro, C.; Lopes, A.R.; Vaz-Moreira, I.; Silva, E.F.; Manaia, C.M.; Nunes, O.C. Wastewater Reuse in Irrigation: A Microbiological Perspective on Implications in Soil Fertility and Human and Environmental Health. Environ. Int. 2015, 75, 117–135.
- Jennings, W.C.; Chern, E.C.; O’Donohue, D.; Kellogg, M.G.; Boehm, A.B. Frequent Detection of a Human Fecal Indicator in the Urban Ocean: Environmental Drivers and Covariation with Enterococci. Environ. Sci. Process. Impacts 2018, 20, 480–492.
- McGinnis, S.; Spencer, S.; Firnstahl, A.; Stokdyk, J.; Borchardt, M.; McCarthy, D.T.; Murphy, H.M. Human Bacteroides and Total Coliforms as Indicators of Recent Combined Sewer Overflows and Rain Events in Urban Creeks. Sci. Total Environ. 2018, 630, 967–976.
- McMinn, B.R.; Ashbolt, N.J.; Korajkic, A. Bacteriophages as Indicators of Faecal Pollution and Enteric Virus Removal. Lett. Appl. Microbiol. 2017, 65, 11–26.
- Figueras, M.J.; Borrego, J.J. New Perspectives in Monitoring Drinking Water Microbial Quality. Int. J. Environ. Res. Public Health 2010, 7, 4179–4202.
- Boehm, A.B.; Van De Werfhorst, L.C.; Griffith, J.F.; Holden, P.A.; Jay, J.A.; Shanks, O.C.; Wang, D.; Weisberg, S.B. Performance of Forty-One Microbial Source Tracking Methods: A Twenty-Seven Lab Evaluation Study. Water Res. 2013, 47, 6812–6828.
- Tillett, B.J.; Sharley, D.; Almeida, M.I.G.; Valenzuela, I.; Hoffmann, A.A.; Pettigrove, V. A Short Work-Flow to Effectively Source Faecal Pollution in Recreational Waters–a Case Study. Sci. Total Environ. 2018, 644, 1503–1510.
- Kirs, M.; Kisand, V.; Wong, M.; Caffaro-Filho, R.A.; Moravcik, P.; Harwood, V.J.; Yoneyama, B.; Fujioka, R.S. Multiple Lines of Evidence to Identify Sewage as the Cause of Water Quality Impairment in an Urbanized Tropical Watershed. Water Res. 2017, 116, 23–33.
- Ahmed, W.; Staley, C.; Sadowsky, M.J.; Gyawali, P.; Sidhu, J.P.; Palmer, A.; Beale, D.J.; Toze, S. Toolbox Approaches Using Molecular Markers and 16S RRNA Gene Amplicon Data Sets for Identification of Fecal Pollution in Surface Water. Appl. Environ. Microbiol. 2015, 81, 7067–7077.
- Dhakar, V.; Geetanjali, A.S. Role of Pepper Mild Mottle Virus as a Tracking Tool for Fecal Pollution in Aquatic Environments. Arch. Microbiol. 2022, 204, 513.
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Kuhn, J.H. Viruses Defined by the Position of the Virosphere within the Replicator Space. Microbiol. Mol. Biol. Rev. 2021, 85, e00120–e00193.
- Gupta, A.; Gupta, R.; Singh, R.L. Microbes and Environment. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Berlin/Heidelberg, Germany, 2017; pp. 43–84.
- Melkikh, A.V. Viruses, Immunity and Evolution. Biosystems 2022, 220, 104761.
- Hernroth, B.E.; Conden-Hansson, A.-C.; Rehnstam-Holm, A.-S.; Girones, R.; Allard, A.K. Environmental Factors Influencing Human Viral Pathogens and Their Potential Indicator Organisms in the Blue Mussel, Mytilus Edulis: The First Scandinavian Report. Appl. Environ. Microbiol. 2002, 68, 4523–4533.
- Kotwal, G.; Cannon, J.L. Environmental Persistence and Transfer of Enteric Viruses. Curr. Opin. Virol. 2014, 4, 37–43.
- Tiwari, R.C.; Dikshit, M.; Mittal, B.; Sharma, V.B. Effects of Water Pollution-A Review Article. J. Ayurveda Integr. Med. Sci. 2022, 7, 63–68.
- Roos, Y.H. Water and Pathogenic Viruses Inactivation—Food Engineering Perspectives. Food Eng. Rev. 2020, 12, 251–267.
- Lanrewaju, A.A.; Enitan-Folami, A.M.; Sabiu, S.; Edokpayi, J.N.; Swalaha, F.M. Global Public Health Implications of Human Exposure to Viral Contaminated Water. Front. Microbiol. 2022, 13, 981896.
- Mihelcic, J.R.; Naughton, C.C.; Verbyla, M.E.; Zhang, Q.; Schweitzer, R.W.; Oakley, S.M.; Wells, E.C.; Whiteford, L.M. The Grandest Challenge of All: The Role of Environmental Engineering to Achieve Sustainability in the World’s Developing Regions. Environ. Eng. Sci. 2017, 34, 16–41.
- Trussell, R.R.; Salveson, A.; Snyder, S.A.; Trussell, R.S.; Gerrity, D.; Pecson, B.M. Potable Reuse: State of the Science Report and Equivalency Criteria for Treatment Trains. WRRF 11-02; WateReuse Research Foundation: Alexandria, VA, USA, 2013; p. 21. Available online: http://www.twdb.texas.gov/publications/reports/contracted_reports/doc/1248321508_Vol2.pdf (accessed on 1 November 2022).
- Carvajal, G.; Roser, D.J.; Sisson, S.A.; Keegan, A.; Khan, S.J. Modelling Pathogen Log10 Reduction Values Achieved by Activated Sludge Treatment Using Naïve and Semi Naïve Bayes Network Models. Water Res. 2015, 85, 304–315.
- Kobayashi, Y.; Ashbolt, N.J.; Davies, E.G.; Liu, Y. Life Cycle Assessment of Decentralized Greywater Treatment Systems with Reuse at Different Scales in Cold Regions. Environ. Int. 2020, 134, 105215.
- Morrison, C.M.; Betancourt, W.Q.; Quintanar, D.R.; Lopez, G.U.; Pepper, I.L.; Gerba, C.P. Potential Indicators of Virus Transport and Removal during Soil Aquifer Treatment of Treated Wastewater Effluent. Water Res. 2020, 177, 115812.
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A Review on Recent Progress in the Detection Methods and Prevalence of Human Enteric Viruses in Water. Water Res. 2018, 135, 168–186.
- Jiang, S.C.; Bischel, H.N.; Goel, R.; Rosso, D.; Sherchan, S.P.; Whiteson, K.L.; Yan, T.; Solo-Gabriele, H.M. Integrating Virus Monitoring Strategies for Safe Non-Potable Water Reuse. Water 2022, 14, 1187.
- Kristanti, R.A.; Hadibarata, T.; Syafrudin, M.; Yılmaz, M.; Abdullah, S. Microbiological Contaminants in Drinking Water: Current Status and Challenges. Water. Air. Soil Pollut. 2022, 233, 299.
- Okoh, A.I.; Sibanda, T.; Gusha, S.S. Inadequately Treated Wastewater as a Source of Human Enteric Viruses in the Environment. Int. J. Environ. Res. Public Health 2010, 7, 2620–2637.
- Bouseettine, R.; Hassou, N.; Bessi, H.; Ennaji, M.M. Waterborne Transmission of Enteric Viruses and Their Impact on Public Health. In Emerging and Reemerging Viral Pathogens; Elsevier: Amsterdam, The Netherlands, 2020; pp. 907–932.
- Liang, G.; Bushman, F.D. The Human Virome: Assembly, Composition and Host Interactions. Nat. Rev. Microbiol. 2021, 19, 514–527.
- Cooper, D.M.; McDonald, J.E.; Malham, S.K.; de Rougemont, A.; Jones, D.L. Seasonal and Spatial Dynamics of Enteric Viruses in Wastewater and in Riverine and Estuarine Receiving Waters. Sci. Total Environ. 2018, 634, 1174–1183.
- Kauppinen, A.; Pitkänen, T.; Miettinen, I.T. Persistent Norovirus Contamination of Groundwater Supplies in Two Waterborne Outbreaks. Food Environ. Virol. 2018, 10, 39–50.
- Walker, D.I.; Adriaenssens, E.M.; McDonald, J.E.; Hillary, L.S.; Malham, S.K.; Jones, D.L. Viral Indicators for Tracking Domestic Wastewater Contamination in the Aquatic Environment. Water Res. 2020, 181, 115926.
- Chatziprodromidou, I.P.; Bellou, M.; Vantarakis, G.; Vantarakis, A. Viral outbreaks linked to fresh produce consumption: A systematic review. J. Appl. Microbiol. 2018, 124, 932–942.
- Al-Ansari, N.; Abbas, N.; Laue, J.; Knutsson, S. Water Scarcity: Problems and Possible Solutions. J. Earth Sci. Geotech. Eng. 2021, 11, 243–312.
- Corpuz, M.V.A.; Buonerba, A.; Vigliotta, G.; Zarra, T.; Ballesteros, F., Jr.; Campiglia, P.; Belgiorno, V.; Korshin, G.; Naddeo, V. Viruses in Wastewater: Occurrence, Abundance and Detection Methods. Sci. Total Environ. 2020, 745, 140910.
- Errani, F.; Ciulli, S.; Mandrioli, L.; Serratore, P.; Volpe, E. Detection of Human and Fish Viruses in Marine Gastropods. Animals 2022, 12, 2122.
- Bányai, K.; Estes, M.K.; Martella, V.; Parashar, U.D. Viral Gastroenteritis. Lancet 2018, 392, 175–186.
- Harper, A.; Vijayakumar, V.; Ouwehand, A.C.; Ter Haar, J.; Obis, D.; Espadaler, J.; Binda, S.; Desiraju, S.; Day, R. Viral Infections, the Microbiome and Probiotics. Front. Cell Infect. Microbiol. 2021, 10, 596166.
- Lopman, B.A.; Steele, D.; Kirkwood, C.D.; Parashar, U.D. The Vast and Varied Global Burden of Norovirus: Prospects for Prevention and Control. PLoS Med. 2016, 13, e1001999.
- Montasser, K.A.; Youssef, M.I.; Ghandour, A.A.; Kamal, M. Infection with Adenovirus, Rotavirus, and Coinfection among Hospitalized Children with Gastroenteritis in an Egyptian University Hospital. J. Med. Virol. 2022, 94, 4950–4958.
- Nour, I.; Hanif, A.; Ryan, M.; Eifan, S. Insights into Gastrointestinal Virome: Etiology and Public Exposure. Water 2021, 13, 2794.
- Torre, P.; Aglitti, A.; Masarone, M.; Persico, M. Viral Hepatitis: Milestones, Unresolved Issues, and Future Goals. World J. Gastroenterol. 2021, 27, 4603.
- King, A.M.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus Taxonomy: Classification and Nomenclature of Viruses. In Virus Taxonomy: Classification and Nomenclature of Viruses; Springer: Berlin/Heidelberg, Germany, 2012; p. 1327.
- Lun, J.H.; Crosbie, N.D.; White, P.A. Genetic Diversity and Quantification of Human Mastadenoviruses in Wastewater from Sydney and Melbourne, Australia. Sci. Total Environ. 2019, 675, 305–312.
- Verani, M.; Casini, B.; Battistini, R.; Pizzi, F.; Rovini, E.; Carducci, A. One-Year Monthly Monitoring of Torque Teno Virus (TTV) in River Water in Italy. Water Sci. Technol. 2006, 54, 191–195.
- De Benedictis, P.; Schultz-Cherry, S.; Burnham, A.; Cattoli, G. Astrovirus Infections in Humans and Animals–Molecular Biology, Genetic Diversity, and Interspecies Transmissions. Infect. Genet. Evol. 2011, 11, 1529–1544.
- Hamad, A.Y.; Mohammed, N.S.; Hasan, A.S. Molecular Detection and Phylogenetic Analysis of Astrovirus and Noroviruses from Sewage Water in Diyala-Iraq. J. Pharm. Negat. Results 2022, 13, 339–347.
- Pérez-Sautu, U.; Sano, D.; Guix, S.; Kasimir, G.; Pintó, R.M.; Bosch, A. Human Norovirus Occurrence and Diversity in the Llobregat River Catchment, Spain. Environ. Microbiol. 2012, 14, 494–502.
- Haramoto, E.; Katayama, H.; Phanuwan, C.; Ohgaki, S. Quantitative Detection of Sapoviruses in Wastewater and River Water in Japan. Lett. Appl. Microbiol. 2008, 46, 408–413.
- Blinkova, O.; Rosario, K.; Li, L.; Kapoor, A.; Slikas, B.; Bernardin, F.; Breitbart, M.; Delwart, E. Frequent Detection of Highly Diverse Variants of Cardiovirus, Cosavirus, Bocavirus, and Circovirus in Sewage Samples Collected in the United States. J. Clin. Microbiol. 2009, 47, 3507–3513.
- Breitbart, M.; Delwart, E.; Rosario, K.; Segalés, J.; Varsani, A.; Consortium, I.R. ICTV Virus Taxonomy Profile: Circoviridae. J. Gen. Virol. 2017, 98, 1997.
- Purdy, M.A.; Harrison, T.J.; Jameel, S.; Meng, X.-J.; Okamoto, H.; Van der Poel, W.H.M.; Smith, D.B.; Consortium, I.R. ICTV Virus Taxonomy Profile: Hepeviridae. J. Gen. Virol. 2017, 98, 2645.
- Hennechart-Collette, C.; Dehan, O.; Laurentie, M.; Fraisse, A.; Martin-Latil, S.; Perelle, S. Method for Detecting Norovirus, Hepatitis A and Hepatitis E Viruses in Tap and Bottled Drinking Water. Int. J. Food Microbiol. 2022, 377, 109757.
- Ahmed, N.I.; Elmahdy, E.M.; Allayh, A.K.; Mohamed, E.-C.B.; Loutfy, S.A.; Barakat, A.; Ali, M.A. Prevalence of Human Polyomavirus and Papillomavirus in Wastewater and in Stool of Egyptian Patients. Egypt. J. Aquat. Biol. Fish. 2019, 23, 29–41.
- Van Doorslaer, K.; Chen, Z.; Bernard, H.-U.; Chan, P.K.; DeSalle, R.; Dillner, J.; Forslund, O.; Haga, T.; McBride, A.A.; Villa, L.L. ICTV Virus Taxonomy Profile: Papillomaviridae. J. Gen. Virol. 2018, 99, 989–990.
- Booranathawornsom, T.; Pombubpa, K.; Tipayamongkholgul, M.; Kittigul, L. Molecular Characterization of Human Bocavirus in Recycled Water and Sewage Sludge in Thailand. Infect. Genet. Evol. 2022, 100, 105276.
- Lodder, W.J.; Rutjes, S.A.; Takumi, K.; de Roda Husman, A.M. Aichi Virus in Sewage and Surface Water, the Netherlands. Emerg. Infect. Dis. 2013, 19, 1222.
- Prevost, B.; Lucas, F.S.; Goncalves, A.; Richard, F.; Moulin, L.; Wurtzer, S. Large Scale Survey of Enteric Viruses in River and Waste Water Underlines the Health Status of the Local Population. Environ. Int. 2015, 79, 42–50.
- El-Senousy, W.M.; Abdel-Moneim, A.; Abdel-Latif, M.; El-Hefnawy, M.H.; Khalil, R.G. Coxsackievirus B4 as a Causative Agent of Diabetes Mellitus Type 1: Is There a Role of Inefficiently Treated Drinking Water and Sewage in Virus Spreading? Food Environ. Virol. 2018, 10, 89–98.
- Hsu, B.-M.; Chen, C.-H.; Wan, M.-T. Genetic Diversity of Epidemic Enterovirus 71 Strains Recovered from Clinical and Environmental Samples in Taiwan. Virus Res. 2007, 126, 69–75.
- Bredykhina, M.; Shtepa, O.; Rezvykh, V.; Paliychuk, O.; Yurchenko, O.; Kovalenko, S.; Hernets, I. Wastewater as an Indicator of Virus Circulation among Population of Dnipropetrovsk Oblast, Ukraine. Online J. Public Health Inform. 2019, 11, e420.
- Matos, A.; Mesquito, J.R.; Goncalves, D.; Abreu-Silva, J.; Luxo, C.; Nascimento, M.S. First Detection and Molecular Characterization of Hepatitis E Virus in Water from Wastewater Treatment Plants in Portugal. Ann. Agric. Environ. Med. 2018, 25, 364–367.
- Moens, U.; Calvignac-Spencer, S.; Lauber, C.; Ramqvist, T.; Feltkamp, M.C.; Daugherty, M.D.; Verschoor, E.J.; Ehlers, B. ICTV Virus Taxonomy Profile: Polyomaviridae. J. Gen. Virol. 2017, 98, 1159–1160.
- Levican, J.; Levican, A.; Ampuero, M.; Gaggero, A. JC Polyomavirus Circulation in One-Year Surveillance in Wastewater in Santiago, Chile. Infect. Genet. Evol. 2019, 71, 151–158.
- Miura, T.; Kadoya, S.; Takino, H.; Sano, D.; Akiba, M. Temporal Variations of Human and Animal Rotavirus A Genotypes in Surface Water Used for Drinking Water Production. Front. Microbiol. 2022, 13, 912147.
- Upfold, N.S.; Luke, G.A.; Knox, C. Occurrence of Human Enteric Viruses in Water Sources and Shellfish: A Focus on Africa. Food Environ. Virol. 2021, 13, 1–31.
- Joshi, M.S.; Lole, K.S.; Barve, U.S.; Salve, D.S.; Ganorkar, N.N.; Chavan, N.A.; Shinde, M.S.; Gopalkrishna, V. Investigation of a Large Waterborne Acute Gastroenteritis Outbreak Caused by Group B Rotavirus in Maharashtra State, India. J. Med. Virol. 2019, 91, 1877–1881.
- Kauppinen, A.; Pitkänen, T.; Al-Hello, H.; Maunula, L.; Hokajärvi, A.-M.; Rimhanen-Finne, R.; Miettinen, I.T. Two Drinking Water Outbreaks Caused by Wastewater Intrusion Including Sapovirus in Finland. Int. J. Environ. Res. Public Health 2019, 16, 4376.
- Naik, S.R.; Aggarwal, R.; Salunke, P.N.; Mehrotra, N.N. A Large Waterborne Viral Hepatitis E Epidemic in Kanpur, India. Bull. World Health Organ. 1992, 70, 597.
- Rachmadi, A.T.; Torrey, J.R.; Kitajima, M. Human Polyomavirus: Advantages and Limitations as a Human-Specific Viral Marker in Aquatic Environments. Water Res. 2016, 105, 456–469.
- Di Bonito, P.; Iaconelli, M.; Gheit, T.; Tommasino, M.; Della Libera, S.; Bonadonna, L.; La Rosa, G. Detection of Oncogenic Viruses in Water Environments by a Luminex-Based Multiplex Platform for High Throughput Screening of Infectious Agents. Water Res. 2017, 123, 549–555.
- Hamza, H.; Hamza, I.A. Oncogenic Papillomavirus and Polyomavirus in Urban Sewage in Egypt. Sci. Total Environ. 2018, 610, 1413–1420.
- Fratini, M.; Di Bonito, P.; La Rosa, G. Oncogenic Papillomavirus and Polyomavirus in Water Environments: Is There a Potential for Waterborne Transmission? Food Environ. Virol. 2014, 6, 1–12.
- Allander, T.; Tammi, M.T.; Eriksson, M.; Bjerkner, A.; Tiveljung-Lindell, A.; Andersson, B. Cloning of a Human Parvovirus by Molecular Screening of Respiratory Tract Samples. Proc. Natl. Acad. Sci. USA 2005, 102, 12891–12896.
- Myrmel, M.; Lange, H.; Rimstad, E. A 1-Year Quantitative Survey of Noro-, Adeno-, Human Boca-, and Hepatitis E Viruses in Raw and Secondarily Treated Sewage from Two Plants in Norway. Food Environ. Virol. 2015, 7, 213–223.
- Adriaenssens, E.M.; McDonald, J.E.; Jones, D.L.; Allison, H.E.; McCarthy, A.J. Tracing the Fate of Wastewater Viruses Reveals Catchment-Scale Virome Diversity and Connectivity. Water Res. 2021, 203, 117568.
- Bibby, K.; Peccia, J. Identification of Viral Pathogen Diversity in Sewage Sludge by Metagenome Analysis. Environ. Sci. Technol. 2013, 47, 1945–1951.
- Lebarbenchon, C.; Yang, M.; Keeler, S.P.; Ramakrishnan, M.A.; Brown, J.D.; Stallknecht, D.E.; Sreevatsan, S. Viral Replication, Persistence in Water and Genetic Characterization of Two Influenza A Viruses Isolated from Surface Lake Water. PLoS ONE 2011, 6, e26566.
- Pachepsky, Y.A.; Allende, A.; Boithias, L.; Cho, K.; Jamieson, R.; Hofstra, N.; Molina, M. Microbial Water Quality: Monitoring and Modeling. J. Environ. Qual. 2018, 47, 931–938.
- Charles, K.J.; Nowicki, S.; Bartram, J.K. A Framework for Monitoring the Safety of Water Services: From Measurements to Security. NPJ Clean Water 2020, 3, 36.
- Ishii, S.; Sadowsky, M.J. Escherichia Coli in the Environment: Implications for Water Quality and Human Health. Microbes Environ. 2008, 23, 101–108.
- Slade, J.S.; Ford, B.J. Discharge to the Environment of Viruses in Wastewater, Sludges, and Aerosols. In Viral Pollution of the Environment; CRC Press: Boca Raton, FL, USA, 2018; pp. 3–16.
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A Review of Methods for the Detection of Pathogenic Microorganisms. Analyst 2019, 144, 396–411.
- Edge, T.A.; Boehm, A.B. Classical and Molecular Methods to Measure Fecal Bacteria. Fecal Bact. 2010, 2, 241–273.
- Sadeghi, S.; Nikaeen, M.; Mohammadi, F.; Nafez, A.H.; Gholipour, S.; Shamsizadeh, Z.; Hadi, M. Microbial Characteristics of Municipal Solid Waste Compost: Occupational and Public Health Risks from Surface Applied Compost. Waste Manag. 2022, 144, 98–105.
- Rock, C.M.; Brassill, N.; Dery, J.L.; Carr, D.; McLain, J.E.; Bright, K.R.; Gerba, C.P. Review of Water Quality Criteria for Water Reuse and Risk-Based Implications for Irrigated Produce under the FDA Food Safety Modernization Act, Produce Safety Rule. Environ. Res. 2019, 172, 616–629.
- Ho, J.Y.; Lavinya, A.A.; Kay, D.S.W.; Lee, C.I.S.; Razmi, A.H.; Walsh, C.L.; Goodson, M.L.; Eswaran, J. Towards an Integrated Approach to Improve the Understanding of the Relationships Between Water-Borne Infections and Health Outcomes: Using Malaysia as a Detailed Case Study. Front. Water 2022, 4, 779860.
- Bisseux, M.; Colombet, J.; Mirand, A.; Roque-Afonso, A.-M.; Abravanel, F.; Izopet, J.; Archimbaud, C.; Peigue-Lafeuille, H.; Debroas, D.; Bailly, J.-L. Monitoring Human Enteric Viruses in Wastewater and Relevance to Infections Encountered in the Clinical Setting: A One-Year Experiment in Central France, 2014 to 2015. Eurosurveillance 2018, 23, 17-00183.
- Sinclair, R.G.; Rose, J.B.; Hashsham, S.A.; Gerba, C.P.; Haas, C.N. Criteria for Selection of Surrogates Used to Study the Fate and Control of Pathogens in the Environment. Appl. Environ. Microbiol. 2012, 78, 1969–1977.
- Jankowski, P.; Gan, J.; Le, T.; McKennitt, M.; Garcia, A.; Yanaç, K.; Yuan, Q.; Uyaguari-Diaz, M. Metagenomic Community Composition and Resistome Analysis in a Full-Scale Cold Climate Wastewater Treatment Plant. Environ. Microb. 2022, 17, 3.
- Savichtcheva, O.; Okabe, S. Alternative Indicators of Fecal Pollution: Relations with Pathogens and Conventional Indicators, Current Methodologies for Direct Pathogen Monitoring and Future Application Perspectives. Water Res. 2006, 40, 2463–2476.
- McCall, C.; Xagoraraki, I. Metagenomic Approaches for Detecting Viral Diversity in Water Environments. J. Environ. Eng. 2019, 148, 04019039.
- Ogorzaly, L.; Bertrand, I.; Paris, M.; Maul, A.; Gantzer, C. Occurrence, Survival, and Persistence of Human Adenoviruses and F-Specific RNA Phages in Raw Groundwater. Appl. Environ. Microbiol. 2010, 76, 8019.
- Enriquez, C.E.; Hurst, C.J.; Gerba, C.P. Survival of the Enteric Adenoviruses 40 and 41 in Tap, Sea, and Waste Water. Water Res. 1995, 29, 2548–2553.
- Rzeżutka, A.; Cook, N. Survival of Human Enteric Viruses in the Environment and Food. FEMS Microbiol. Rev. 2004, 28, 441–453.
- Pinon, A.; Vialette, M. Survival of Viruses in Water. Intervirology 2018, 61, 214–222.
- Davidson, P.C.; Kuhlenschmidt, T.B.; Bhattarai, R.; Kalita, P.K.; Kuhlenschmidt, M.S. Investigation of Rotavirus Survival in Different Soil Fractions and Temperature Conditions. J. Environ. Prot. 2013, 4, 34107.
- Raphael, R.A.; Sattar, S.A.; Springthorpe, V.S. Long-Term Survival of Human Rotavirus in Raw and Treated River Water. Can. J. Microbiol. 1985, 31, 124–128.
- Seitz, S.R.; Leon, J.S.; Schwab, K.J.; Lyon, G.M.; Dowd, M.; McDaniels, M.; Abdulhafid, G.; Fernandez, M.L.; Lindesmith, L.C.; Baric, R.S. Norovirus Infectivity in Humans and Persistence in Water. Appl. Environ. Microbiol. 2011, 77, 6884.
- Ngazoa, E.S.; Fliss, I.; Jean, J. Quantitative Study of Persistence of Human Norovirus Genome in Water Using TaqMan Real-Time RT-PCR. J. Appl. Microbiol. 2008, 104, 707–715.
- Callahan, K.M.; Taylor, D.J.; Sobsey, M.D. Comparative Survival of Hepatitis A Virus, Poliovirus and Indicator Viruses in Geographically Diverse Seawaters. Water Sci. Technol. 1995, 31, 189–193.
- Crance, J.M.; Gantzer, C.; Schwartzbrod, L.; Deloince, R. Effect of Temperature on the Survival of Hepatitis A Virus and Its Capsidal Antigen in Synthetic Seawater. Environ. Toxicol. Water Qual. Int. J. 1998, 13, 89–92.
- Leblanc, D.; Gagné, M.-J.; Poitras, É.; Brassard, J. Persistence of Murine Norovirus, Bovine Rotavirus, and Hepatitis A Virus on Stainless Steel Surfaces, in Spring Water, and on Blueberries. Food Microbiol. 2019, 84, 103257.
- Abad, F.X.; Pintó, R.M.; Villena, C.; Gajardo, R.; Bosch, A. Astrovirus Survival in Drinking Water. Appl. Environ. Microbiol. 1997, 63, 3119–3122.
- Morsy El-Senousy, W. Suitability of Some Viruses as Indices of Viral Pollution of Water. Egypt. J. Aquat. Biol. Fish. 2021, 25, 1049–1084.
- Franco, E.; Meleleo, C.; Serino, L.; Sorbara, D.; Zaratti, L. Hepatitis A: Epidemiology and Prevention in Developing Countries. World J. Hepatol. 2012, 4, 68.
- Ahmed, W.; Bivins, A.; Payyappat, S.; Cassidy, M.; Harrison, N.; Besley, C. Distribution of Human Fecal Marker Genes and Their Association with Pathogenic Viruses in Untreated Wastewater Determined Using Quantitative PCR. Water Res. 2022, 226, 119093.
- Canh, V.D.; Lien, N.T.; Nga, T.T.V. Evaluation of the Suitability of Pepper Mild Mottle Virus (PMMoV) as an Indicator Virus for Water Safety and Quality. J. Sci. Technol. Civ. Eng. 2022, 16, 76–88.
More
