Human transcriptomes are more divergent than genes and contribute to the sophistication of life. This divergence is derived from various isoforms arising from alternative splicing. In addition, alternative splicing regulated by spliceosomal factors and RNA structures, such as the RNA G-quadruplex, is important not only for isoform diversity but also for regulating gene expression. Therefore, abnormal splicing leads to serious diseases such as cancer and neurodegenerative disorders.We describe the regulation of divergent transcriptomes using alternative mRNA splicing.
- alternative mRNA splicing
- cancer
- neurodegenerative disorder
- splicing inhibitor
1. Introduction
In eukaryotic cells, protein-coding genes are transcribed as pre-messenger RNA (mRNA) in the nucleus and pre-mRNA undergoes several RNA processing steps, such as 5′-capping, splicing, and 3′-end processing. These gene expression processes are tightly coordinated with each other to achieve efficient and accurate gene expression [1]. After the mRNA processing steps, the mature mRNA is exported from the nucleus to the cytoplasm for translation. Human transcriptomes are more divergent than genes. This divergence is derived from various isoforms arising from alternative splicing, which is an essential biological process for considerable proteomic diversity and complexity despite the relatively limited number of human genes [2]. Furthermore, alternative splicing has important roles not only for expressing proteins through transcript diversity but also for regulating gene expression. Transcripts from most human protein-coding genes undergo one or more forms of alternative splicing. Alternatively spliced isoforms vary greatly from tissue to tissue [3]. Recent comprehensive analysis suggested that more than 40% of genes express multiple isoforms in a single tissue [4]. In particular, many isoforms containing alternative exons are expressed in neurons; thus, the brain displays the most complex pattern of alternative splicing [5]. Alternative splicing contributes to cell differentiation and lineage determination, tissue identity acquisition and maintenance, and organ development [6][7][8]. Thus, alternative splicing is considered to be a key mechanism for regulating gene expression networks, as well as for human diversity or sophistication.
2. Data
The spliceosome, a multimegadalton ribonucleoprotein (RNP) complex comprising five snRNPs and numerous proteins, carries out splicing of pre-mRNA molecules to remove introns, then conjoins exons in different arrangements that potentially encode alternative protein isoforms [9]. Spliceosome complexes are assembled at the splice sites in a pre-mRNA transcript, involving a stepwise assembly pathway of the U1, U2, and U4/5/6 snRNP spliceosome subunits. The U1 snRNP binds the 5′ splice site (5′ ss), and splicing factor 1 (SF1) and U2 snRNP auxiliary factor (U2AF) 1/2 bind the branch point (BP), polypyrimidine tract, and the 3′ ss, respectively (
). After binding, the U2 snRNP containing the splicing factor 3A (SF3A) and splicing factor 3B (SF3B) subcomplex stably associates with BP, following the engagement of U4/U6 and U5 snRNPs in the form of a tri-snRNP particle. This leads to the destabilization of the U1 and U4 snRNPs. These conformational and compositional rearrangements of spliceosomal components result in the activated spliceosome; this emergence triggers two sequential transesterification reactions to produce the spliced-mRNA. Additional interactions that contribute to the recognition of intron–exon boundaries and/or the spliceosome assembly are mediated by elements of the cis-acting exonic-splicing enhancer (ESE) and intronic-splicing enhancer, and exonic-splicing silencer (ESS) and intronic-splicing silencer, which are recognized by auxiliary splicing factors, including the Ser/Arg-rich (SR) proteins (hereafter described as SRSFs) and heterogeneous nuclear ribonucleoproteins (hnRNPs) [10]. Strict recognition of the splicing site by these factors enables individual alternative splicing.
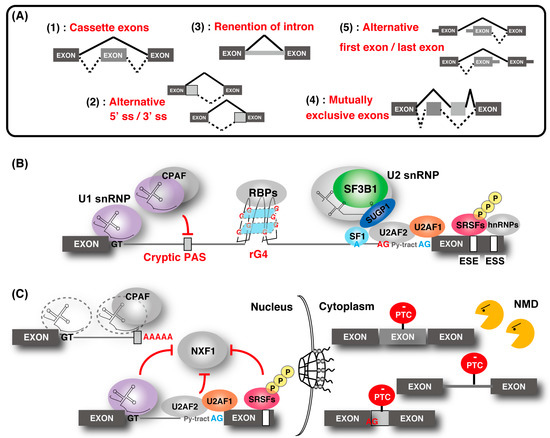
Regulation of gene expression through alternative splicing. (
) Five types of alternative splicing events. "Cassette exons" means inclusion or skipping of an exon. (
) Outline of the alternative splicing control by various factors. The U1 ribonucleoprotein (snRNP) function to recognize the 5′ splice site (ss) and the cryptic polyadenylation signals (PASs) with cleavage and polyadenylation factors (CPAFs). Splicing factor 1 (SF1), U2 snRNP auxiliary factor 1 (U2AF1), and U2AF2 bind the branch point (BP; shown as a blue “A”), polypyrimidine tract (shown as “Py-tract”), and 3′ ss (shown as a blue “AG”), respectively. U2 snRNP containing splicing factor 3B1 (SF3B1), SURP and G-patch domain containing 1 (SUGP1) displaces SF1 and binds to the BP. SR protein families and hnRNPs recognize the exonic-splicing enhancer (ESE) and exonic-splicing silencer(ESS) elements, respectively, and contribute to the recognition of the splice site and the spliceosome assembly by splicing factors. The cryptic 3′ ss (shown as a red “AG”) recognition is repressed by hnRNP. RNA G-quadruplex (rG4) structures affect splicing by acting as RNA-binding protein motifs. (
) Association of alternative splicing and gene regulation. Without U1 snRNP, the CPAF recognizes intronic PASs and generates short transcripts due to premature cleavage and polyadenylation. Some intron-containing transcripts are associated with U1 snRNP, U2AF2, and SR proteins, and they are tethered in the nucleus. Intron-containing transcripts, which are exported to the cytoplasm, often contain a premature termination codon (PTC) and are eliminated by nonsense-mediated messenger RNA (mRNA) decay (NMD). Similarly, other alternative and cryptic-spliced transcripts with PTC are also degraded by NMD.
Splicing contributes to precise gene regulation in connection with other forms of processing in the nucleus. For example, splicing of first introns feeds back to transcription elongation and the efficiency of last intron removal affects cleavage and polyadenylation of mRNAs [11][12][13]. Coupling mRNA splicing to mRNA export ensures efficient nuclear export of mature mRNPs for translation in the cytoplasm mediated by the evolutionarily conserved transcription and export (TREX) complex [1][14]. The TREX complex is recruited to mRNA in a splicing-dependent manner via splicing factor and UAP56, and it triggers the association of nuclear RNA export factor 1 (NXF1), which is a final mRNA export factor, onto the export competent mRNA. Recently, a molecular mechanism that suppresses the recruitment of NXF1 to incompletely spliced mRNAs was partly demonstrated [15][16][17]. In addition, gene regulation through nonsense-mediated mRNA decay (NMD) was shown [18][19][20][21][22][23][24].The aberrant transcript with the pre-mature termination codon, derived from abnormal splicing, is removed by NMD.
References
- Carey, K.T.; Wickramasinghe, V.O. Regulatory Potential of the RNA Processing Machinery: Implications for Human Disease. Trends Genet. 2018, 34, 279–290.
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflügers Arch. Eur. J. Physiol. 2018, 470, 995–1016.
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476.
- Sterne-Weiler, T.; Weatheritt, R.J.; Best, A.J.; Ha, K.C.H.; Blencowe, B.J. Efficient and Accurate Quantitative Profiling of Alternative Splicing Patterns of Any Complexity on a Laptop. Cell 2018, 72, 187–200.
- Porter, R.S.; Jaamour, F.; Iwase, S. Neuron-specific alternative splicing of transcriptional machineries: Implications for neurodevelopmental disorders. Cell. Neurosci. 2018, 87, 35–45.
- Kalsotra, A.; Xiao, X.; Ward, A.J.; Castle, J.C.; Johnson, J.M.; Burge, C.B.; Cooper, T.A. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Natl. Acad. Sci. USA 2008, 105, 20333–20338.
- Dillman, A.A.; Hauser, D.N.; Gibbs, J.R.; Nalls, M.A.; McCoy, M.K.; Rudenko, I.N.; Galter, D.; Cookson, M.R. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Neurosci. 2013, 16, 499–506.
- Bhate, A.; Parker, D.J.; Bebee, T.W.; Ahn, J.; Arif, W.; Rashan, E.H.; Chorghade, S.; Chau, A.; Lee, J.-H.; Anakk, S.; et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Commun. 2015, 6, 8768.
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, 1–2.
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Rev. Genet. 2016, 17, 19–32.
- Bieberstein, N.I.; Oesterreich, F.C.; Straube, K.; Neugebauer, K.M. First exon length controls active chromatin signatures and transcription. Cell Rep.2012, 2, 62–68.
- Martinson, H.G. An active role for splicing in 3′-end formation. Wiley Interdiscip. Rev. RNA 2011, 2, 459–470.
- Movassat, M.; Crabb, T.L.; Busch, A.; Yao, C.; Reynolds, D.J.; Shi, Y.; Hertel, K.J. Coupling between alternative polyadenylation and alternative splicing is limited to terminal introns. RNA Biol. 2016, 13, 646–655.
- Heath, C.G.; Viphakone, N.; Wilson, S.A. The role of TREX in gene expression and disease. J. 2016, 473, 2911–2935.
- Müller-McNicoll, M.; Botti, V.; de Jesus Domingues, A.M.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566.
- Botti, V.; McNicoll, F.; Steiner, M.C.; Richter, F.M.; Solovyeva, A.; Wegener, M.; Schwich, O.D.; Poser, I.; Zarnack, K.; Wittig, I.; et al. Cellular differentiation state modulates the mRNA export activity of SR proteins. Cell Biol. 2017, 216, 1993–2009.
- Wegener, M.; Müller-McNicoll, M. Nuclear retention of mRNAs—Quality control, gene regulation and human disease. Cell Dev. Biol. 2018, 79, 131–142.
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. Cell Sci. 2016, 129, 461–467.
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Rev. Mol. Cell Biol.2019, 20, 406–420.
- Jumaa, H.; Nielsen, P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997, 16, 5077–5085.
- Lareau, L.F.; Inada, M.; Green, R.E.; Wengrod, J.C.; Brenner, S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 2007, 446, 926–929.
- Ni, J.Z.; Grate, L.; Donohue, J.P.; Preston, C.; Nobida, N.; O’Brien, G.; Shiue, L.; Clark, T.A.; Blume, J.E.; Ares, M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007, 21, 708–718.
- Rossbach, O.; Hung, L.-H.; Schreiner, S.; Grishina, I.; Heiner, M.; Hui, J.; Bindereif, A. Auto- and Cross-Regulation of the hnRNP L Proteins by Alternative Splicing. Cell. Biol. 2009, 29, 1442–1451.
- McGlincy, N.J.; Tan, L.Y.; Paul, N.; Zavolan, M.; Lilley, K.S.; Smith, C.W.J. Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics 2010, 11, 1–19.
- Anczukow, O.; Krainer, A.R. Splicing-factor alterations in cancers. RNA 2016, 22, 1285–1301.
Mis-regulation of complicated alternative splicing is associated with cancers, and abnormal expression or mutations in splicing factors contribute to tumorigenesis and neurodegenerative disorders [25]. Recently, various compounds with splicing inhibitor activity were established. These chemical compounds are expected to act not only as a biological tool to investigate complicated splicing processes but also for anti-cancer drugs targeted to the splicing machinery. Food-derived compounds having similar functions were also identified and are receiving attention.
References
- Carey, K.T.; Wickramasinghe, V.O. Regulatory Potential of the RNA Processing Machinery: Implications for Human Disease. Trends Genet. 2018, 34, 279–290.
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflügers Arch. Eur. J. Physiol. 2018, 470, 995–1016.
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476.
- Sterne-Weiler, T.; Weatheritt, R.J.; Best, A.J.; Ha, K.C.H.; Blencowe, B.J. Efficient and Accurate Quantitative Profiling of Alternative Splicing Patterns of Any Complexity on a Laptop. Cell 2018, 72, 187–200.
- Porter, R.S.; Jaamour, F.; Iwase, S. Neuron-specific alternative splicing of transcriptional machineries: Implications for neurodevelopmental disorders. Cell. Neurosci. 2018, 87, 35–45.
- Kalsotra, A.; Xiao, X.; Ward, A.J.; Castle, J.C.; Johnson, J.M.; Burge, C.B.; Cooper, T.A. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Natl. Acad. Sci. USA 2008, 105, 20333–20338.
- Dillman, A.A.; Hauser, D.N.; Gibbs, J.R.; Nalls, M.A.; McCoy, M.K.; Rudenko, I.N.; Galter, D.; Cookson, M.R. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Neurosci. 2013, 16, 499–506.
- Bhate, A.; Parker, D.J.; Bebee, T.W.; Ahn, J.; Arif, W.; Rashan, E.H.; Chorghade, S.; Chau, A.; Lee, J.-H.; Anakk, S.; et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Commun. 2015, 6, 8768.
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, 1–2.
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Rev. Genet. 2016, 17, 19–32.
- Bieberstein, N.I.; Oesterreich, F.C.; Straube, K.; Neugebauer, K.M. First exon length controls active chromatin signatures and transcription. Cell Rep.2012, 2, 62–68.
- Martinson, H.G. An active role for splicing in 3′-end formation. Wiley Interdiscip. Rev. RNA 2011, 2, 459–470.
- Movassat, M.; Crabb, T.L.; Busch, A.; Yao, C.; Reynolds, D.J.; Shi, Y.; Hertel, K.J. Coupling between alternative polyadenylation and alternative splicing is limited to terminal introns. RNA Biol. 2016, 13, 646–655.
- Heath, C.G.; Viphakone, N.; Wilson, S.A. The role of TREX in gene expression and disease. J. 2016, 473, 2911–2935.
- Müller-McNicoll, M.; Botti, V.; de Jesus Domingues, A.M.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566.
- Botti, V.; McNicoll, F.; Steiner, M.C.; Richter, F.M.; Solovyeva, A.; Wegener, M.; Schwich, O.D.; Poser, I.; Zarnack, K.; Wittig, I.; et al. Cellular differentiation state modulates the mRNA export activity of SR proteins. Cell Biol. 2017, 216, 1993–2009.
- Wegener, M.; Müller-McNicoll, M. Nuclear retention of mRNAs—Quality control, gene regulation and human disease. Cell Dev. Biol. 2018, 79, 131–142.
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. Cell Sci. 2016, 129, 461–467.
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Rev. Mol. Cell Biol.2019, 20, 406–420.
- Jumaa, H.; Nielsen, P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997, 16, 5077–5085.
- Lareau, L.F.; Inada, M.; Green, R.E.; Wengrod, J.C.; Brenner, S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 2007, 446, 926–929.
- Ni, J.Z.; Grate, L.; Donohue, J.P.; Preston, C.; Nobida, N.; O’Brien, G.; Shiue, L.; Clark, T.A.; Blume, J.E.; Ares, M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007, 21, 708–718.
- Rossbach, O.; Hung, L.-H.; Schreiner, S.; Grishina, I.; Heiner, M.; Hui, J.; Bindereif, A. Auto- and Cross-Regulation of the hnRNP L Proteins by Alternative Splicing. Cell. Biol. 2009, 29, 1442–1451.
- McGlincy, N.J.; Tan, L.Y.; Paul, N.; Zavolan, M.; Lilley, K.S.; Smith, C.W.J. Expression Proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics 2010, 11, 1–19.
- Anczukow, O.; Krainer, A.R. Splicing-factor alterations in cancers. RNA 2016, 22, 1285–1301.
