You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Elisabeth Jamet.
Cell walls are an extracellular compartment specific to plant cells, which is not found in animal cells. Their composition varies between cell types, plant species, and physiological states. They are composed of a great diversity of polymers,
e.g.
, polysaccharides, proteins, and lignins. Cell wall proteins (CWPs) are major players involved in the plasticity of cell walls which support cell growth and differentiation, as well as adaptation to environmental changes. In order to reach the extracellular space, CWPs are transported through the secretory pathway where they may undergo post-translational modifications, including
N
-glycosylations on the Asn residues in specific motifs (Asn-X-Ser/Thr-X, with X≠Pro).
- cell wall
- cell wall protein
- Concanavalin A affinity chromatography
- glycopeptides
1. Introduction
Glycosylations are major post-translational modifications (PTMs) observed in plant cell wall proteins (CWPs), and they occur in the secretory pathway. There are of three types: N-glycosylation, O-glycosylation, and glypiation (for a review, see [1]). N-glycosylation consists of the grafting of an oligosaccharide onto a particular amino acid, asparagine (Asn), in the context of a specific motif, Asn-X-Ser/Thr-X (with X≠Pro). Since it is short (only four amino acids), this motif is classified among the motifs with a high probability of occurrence. It means that the chance to find it in protein sequences is high even if the protein does not travel through the secretion pathway where N-glycans are synthesized and grafted. O-glycosylation occurs on the hydroxyl groups of Ser and hydroxyproline (Hyp) residues located in particular amino acid motifs, especially in the structural proteins of the hydroxyproline-rich glycoprotein families (HRGPs) [3,4,5,6,7][1][2][3][4][5]. Glypiation consists of the addition of a glycosylphosphatidylinositol (GPI) being anchored at the C-terminus of proteins and allows the anchoring of proteins at the external side of the plasma membrane (for a review, see [8]). All these PTMs are crucial for the structure and the function of CWPs. This review will focus on N-glycosylation which affects a great proportion of CWPs.
In plants, there are various types of N-glycans [9][7]. The biosynthesis of N-glycans has already been extensively reviewed (see for example, [9,10,11,12,13,14][7][8][9][10][11][12]), and it will not be described in detail here. Briefly, the N-glycosylation process starts in the endoplasmic reticulum (ER) by the co-translational transfer of an oligosaccharide precursor of the high-mannose type, Glc(3)Man(9)GlcNAc(2) (Figure 1), onto the Asn residues of the N-glycosylation motifs of proteins. Then, these N-glycans undergo several maturation steps consisting of the removal and the subsequent addition of sugar residues in the ER and the Golgi apparatus by glycosidases and glycosyl transferases, respectively; this gives rise to complex N-glycans with Lewis epitopes. The presence of a core α(1,3)-Fuc and a β(1,2)-Xyl is specific to plant N-glycans, compared to mammal N-glycans [9][7]. Overall, different types of N-glycans are generated from complex N-glycans, such as hybrid and paucimannose N-glycans (Figure 1).

Figure 1. Some examples of N-glycan structures grafted onto Asn-X-Ser/Thr motifs (with X≠Pro) of plant N-glycoproteins. They all share a core Man3GlcNAc2 structure. The core structure is substituted by two to six Man residues in high-mannose N-glycans. Complex N-glycans comprise a β-1,2-linked Xyl, an α-1,3-linked Fuc residue, and/or one or two β-1,2-linked GlcNAc residues bound to the core. Hybrid N-glycans share common features with high-mannose and complex N-glycans. Paucimannose N-glycans do not exhibit terminal GlcNAc residues. Some complex N-glycans also comprise one out of the two terminal α-1,4-Fuc and β-1,3-Gal residues which form Lewis a epitopes.
The structure of N-glycans has evolved in the green lineage [10,11][8][9]. In green algae, other types of N-glycans have been found. In the microalgae Chlorella vulgaris, N-glycans are mostly of the high mannose/oligomannose-type, and they are O-methylated at their non-reducing termini [15][13]. In the Chlamydomonas rheinardtii green alga, the main N-glycans are of three types: oligomannose N-glycans represent about 70% of the N-glycan population, and complex N-glycans with one or two pentose residues account for 14.1 and 16.6%, respectively [16][14]. In Penium margaritaceum, a charophycean green alga assumed to be sister to land plants, the same N-glycans as those of flowering plants are found [17][15]. In the bryophyte Marchantia polymorpha, preliminary results indicate the presence of complex N-glycans with two pentose residues (H Kolkas et al., unpublished results). In the moss Physcomitrella patens, the same types of N-glycans as in flowering plants have been characterized [18][16]. These results suggest that the whole machinery of N-glycosylation has been established very early in the green lineage and conserved along the evolution.
The importance of N-glycosylation has been stressed by the study of N-glycosylation mutants. The severity of their phenotypes depends on which step of N-glycan biosynthesis is impaired. The more severe phenotypes are observed when the mutation affects the early steps of N-glycan biosynthesis in the ER, especially the biosynthesis of lipid-linked oligosaccharide (LLO) precursors or the multi-subunit oligosaccharyl transferase (OST) complex. Some mutations can be lethal (for reviews, see [13,19][11][17]). At later stages of N-glycan biosynthesis, mutations can affect post-seedling development, as in the Oryza sativa N-acetylglucosaminyltransferase 1 (gnt1) mutant, but still allow callus development [20][18]. The gnt1 mutant is impaired with the addition of the β(1,2)-linked GlcNAc to the core Man(5)GlcNac(2) which is a prerequisite for the synthesis of complex and hybrid N-glycans. On the contrary, when the mutation affects a late stage of N-glycan biosynthesis, such as the grafting of α(1,3)-Fuc of β(1,2)-Xyl by the core xylosyl transferase (XYLT) or fucosyl transferases (FUT11 and FUT12) in A. thaliana, no developmental phenotype has been observed [21][19].
2. An Overview of the Present N-glycoproteomics Studies
A selection of articles devoted to N-glycoproteomics is presented in Table 1. Several plants have been studied: Marchantia polymorpha [52][20] as an early divergent plant; Zea mays [34][21], Triticum aestivum [33][22] and Brachypodium distachyon [53][23] as monocot plants; and Fagopyrum tataricum [38][24], Solanum lycopersicum [32[25][26][27],37,54], Gossypium hirsutum [55][28], Camellia sinensis [56][29], Arabidopsis thaliana [22,24,36,43,57,58][30][31][32][33][34][35] and Brassica oleracea [23][36] as dicot plants. N-glycoproteomes have been described in different organs: whole thalli corresponding to the haploid gametophytic stage of M. polymorpha [52][20]; seedlings [59][37]; aerial organs for flowering plants, such as actively growing hypocotyls of etiolated seedlings [24][31]; leaves [33,34,43,56][21][22][29][33]; stems [22,43][30][33]; inflorescences [58][35]; fruit pericarp [32,37,54][25][26][27]; and seeds [38,55][24][28]. The N-glycoproteome of the xylem sap of B. oleracea has also been analyzed, and looks very similar to a cell wall proteome [23][36]. Additionally, in a few studies, the N-glycans released by PNGase A, which cleaves N-glycans regardless of the presence of Xyl of Fuc residues, have been analyzed [35,42][38][39].Table 1. A selection of N-glycoproteomes.
| Plant/Green Algae Species |
Organ | Strategy 1 | Size 2 | Predicted CWPs 3 | Reference |
|---|---|---|---|---|---|
| Marchantia polymorpha | thallus 4 | 3 | 249 (92.0%) | 221 (88.8%) | [52][20] |
| Zea mays | seedling leaf | 5/7 | 476 (100%) | 307 (64.5%) | [34][21] |
| Triticum aestivum | seedling leaf | 6/8 | 312 (100%) | 236 (75.6%) | [33][22] |
| Brachypodium distachyon | seedling leaf | 5/7 | 35 (100%) | 28 (80.0%) | [53][23] |
| Fagopyrum tataricum | seed | 5/8 | 285 (100%) | nd | [38][24] |
| Solanum lycopersicum | fruit (pericarp) 4 | 5/8 | 363 (96.7%) | 202 (55.6%) | [37][26] |
| fruit (pericarp) 4 | 1 | 108 (97.2%) | 101 (93.5%) | [54][27] | |
| Gossypium hirsutum | seed(fiber cells) 4 | 1 | 199 (91.5%) | 114 (57.3%) | [55][28] |
| Camellia sinensis | leaf 4 | 5/8 | 382 (97.9%) | 267 (69.9%) | [56][29] |
| Arabidopsis thaliana | hypocotyl (etiolated) 4 | 3 | 127 (91.3%) | 123 (96.9%) | [24][31] |
| seedling | 5/8 | 912 (nd) | nd | [59][37] | |
| seedling/stem/floret | 5/8 | 538 (100%) | 343(64%) | [43][33] | |
| seedling and leaf4 | 9 | 173 (84.0%) | 135 (78.0%) | [36][32] | |
| leaf 4 | 3 | 62 (98.4%) | 58 (93.5%) | [57][34] | |
| stem 4 | 1 | 98 (100%) | 88 (89.8%) | [22][30] | |
| inflorescence 4 | 5/7 | 265 (96.6%) | 190 (71.7%) | [58][35] | |
| Brassica oleracea | xylem sap 4 | 2 | 75 (94.7%) | 74 (98.7%) | [23][36] |
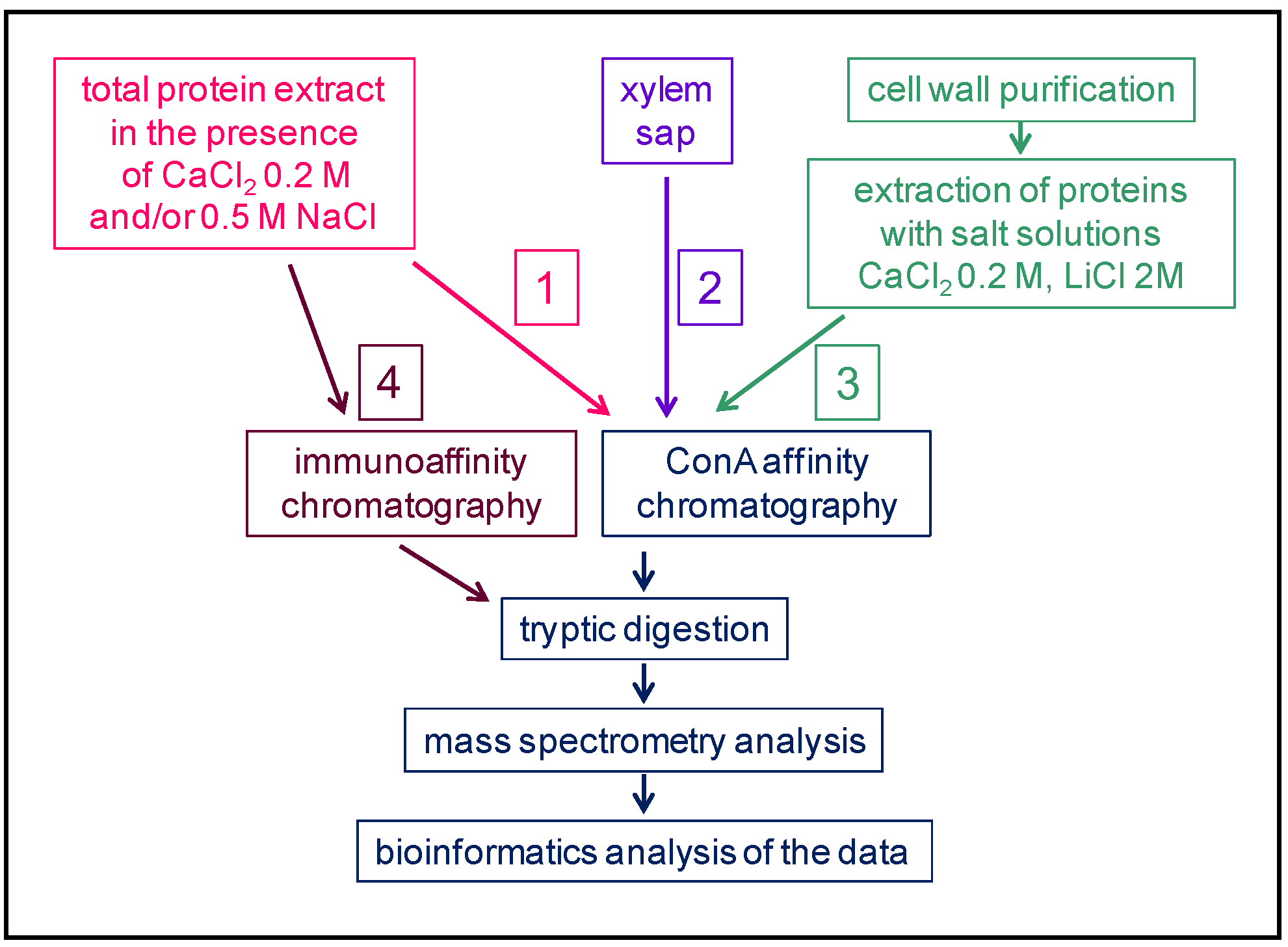
Figure 2. Comparison between cell wall proteomes (red) and N-glycoproteomes (green) characterized from the same plant material. In each case, the overall coverage of the cell wall proteome increases by combining the two approaches.
3. The Heterogeneity of Plant N-glycans
All the N-glycans studied are grafted on an Asn amino acid residue, but their monosaccharide composition is variable. In Ma et al. [59][37], 161 possible structures have been collected in a library to allow the identification of N-glycopeptides and N-glycans in A. thaliana seedlings using MS. The diversity of N-glycans grafted on a given protein is well-illustrated in the case of the A. thaliana α-xylosidase (AtXYL1, At1g68560) belonging to the glycoside hydrolase family 31 [62][42]. Eight N-glycosylation sites could be predicted using the Prosite motif (PS00001) on AtXYL1. Among them, five were experimentally found to be occupied [43][33], and there is no information about the others. Altogether, 19 different N-glycans have been identified in AtXYL1 and 196 N-glycan occurrences have been observed (Figure 3). These N-glycans belong to five classes of N-glycans: A single high mannose glycan is found, as well as one complex N-glycan with a Lewis a epitope. Otherwise, all the identified N-glycans are of the complex-, hybrid-, or paucimannose-type. Major N-glycans could be identified in each class: HexNAc(4)Hex(3)Fuc(1)Pent(1) for complex-type (46 occurrences), HexNAc(3)Hex(3)Pent(1) and HexNAc(3)Hex(3)Fuc(1)Pent(1) for hybrid-type (20 and 31 occurrences, respectively), and HexNAc(2)Hex(3)Fuc(1)Pent(1) for paucimannose-type (46 occurrences).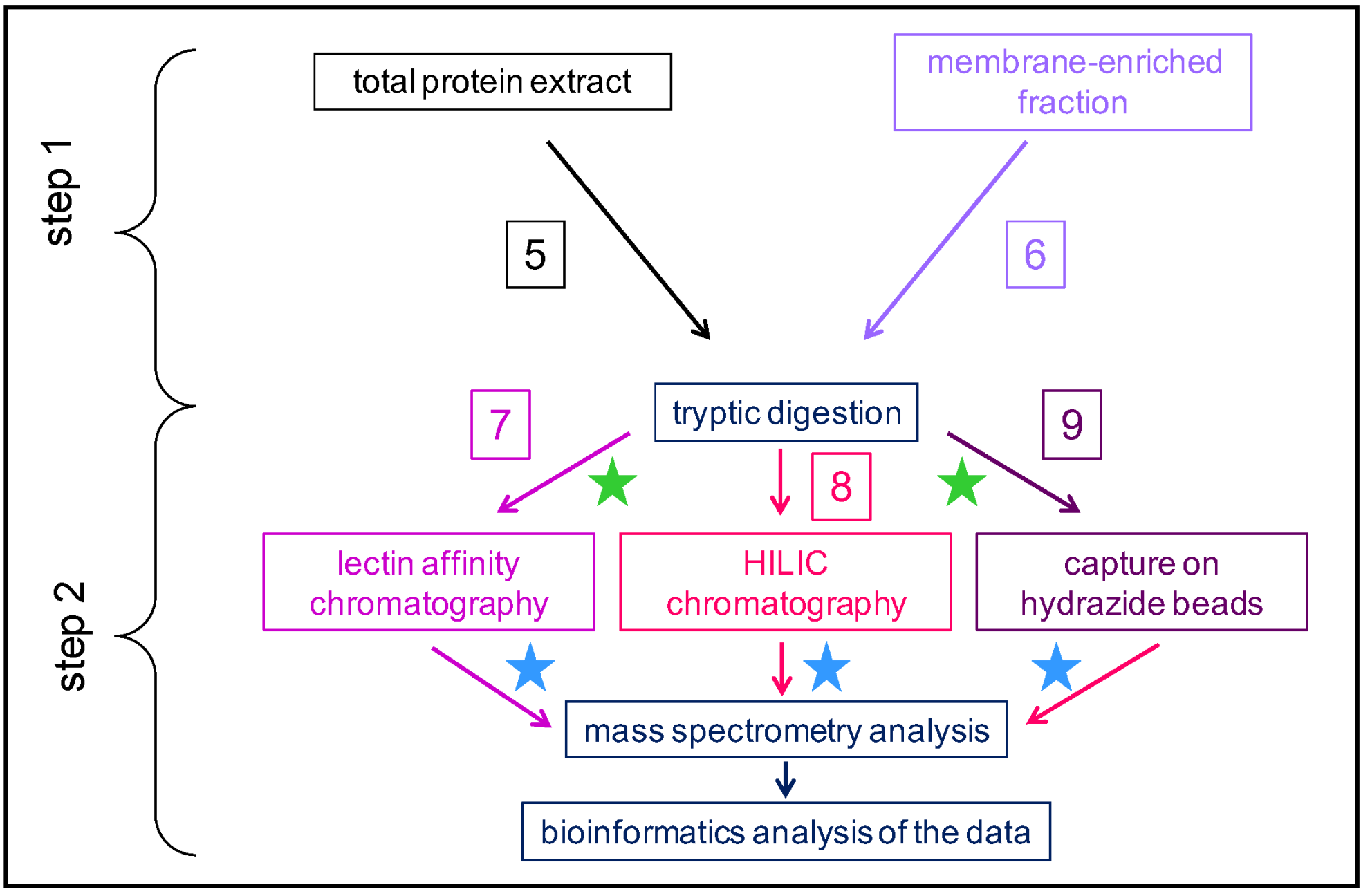
Figure 3. Diversity of N-glycans on a given protein. This figure uses the MS experimental results related to the At1g68560 protein identified in the A. thaliana rosettes N-glycoproteome described in [43][33]. At1g68560 exhibits eight predicted N-glycosylation sites, out of which five are shown to be occupied by N-glycans. Nineteen different N-glycans are found, including a single high mannose-type (light blue), a 53 complex-type (dark blue), a 59 hybrid-type (red), an 82 paucimannose-type (green), and a single complex-type with a Lewis a epitope.
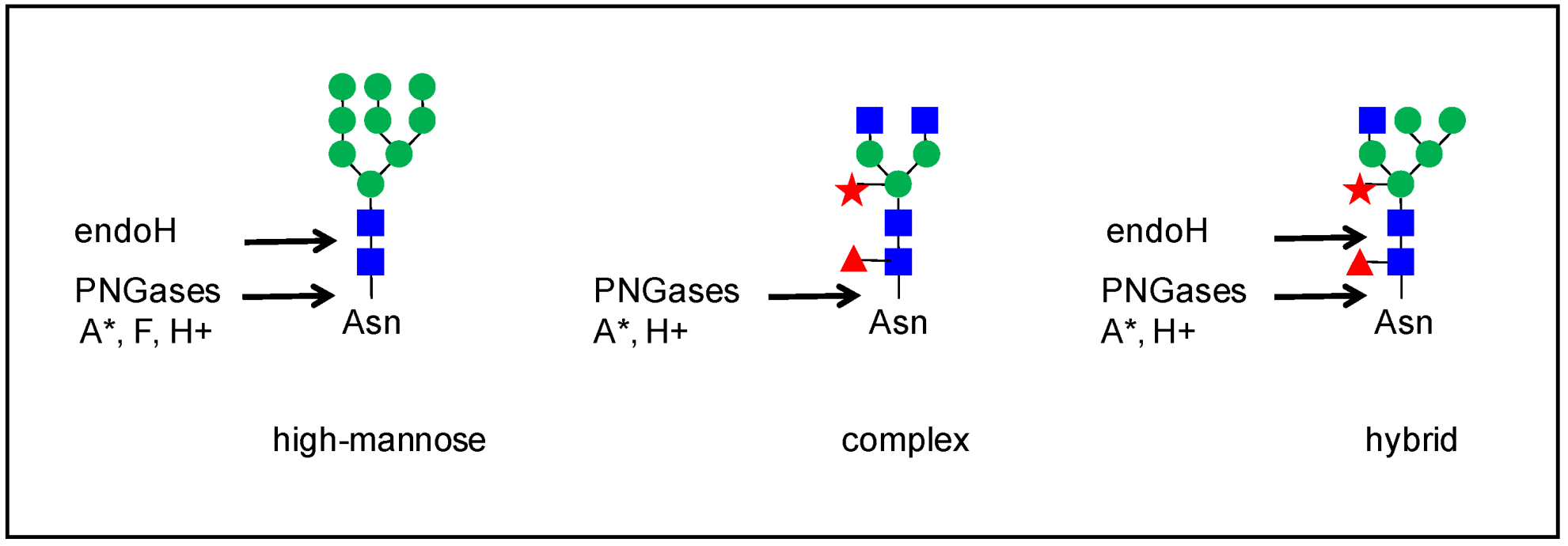
Figure 4. Microheterogeneity of N-glycosylation. This figure uses the MS experimental results related to the At1g68560 protein identified in the A. thaliana rosettes N-glycoproteome described in [43][33]. (A). Types of N-glycans. Each N-glycan type is associated with a symbol. (B). Description of the eight predicted N-glycopeptides in the amino acid sequence of At1g68560. The Asn (N) residue of the underlined consensus N-glycosylation site (PS00001) is in pink, and the tryptic Lys (K) and Arg (R) recognition sites in light blue. In each case, the consensus N-glycosylation motif is underlined. N-glycans have been found for six out of them. (C). Distribution of the different types of N-glycans in one of the glycopeptides, as indicated by an arrow in B. The green area corresponds to the major N-glycan type in each case, whereas the blue area corresponds to the other types of N-glycans: the high mannose-type is represented once by HexNAc(2)Hex(9) (diamond); the major complex-type is HexNAc(4)Hex(3)Fuc(1)Pent(1) (star); the major hybrid-type is HexNAc(3)Hex(3)Fuc(1)Pent(1) (circle); and the major paucimannose-type is HexNAc(2)Hex(3)Fuc(1)Pent(1) (square).
References
- Nguema-Ona, E.; Vicré-Gibouin, M.; Gotté, M.; Plancot, B.; Lerouge, P.; Bardor, M.; Driouich, A. Cell wall O-glycoproteins and N-glycoproteins: Aspects of biosynthesis and function. Front. Plant Sci. 2014, 5, 499.
- Kieliszewski, M.J. The latest hype on Hyp-O-glycosylation codes. Phytochemistry 2001, 57, 319–323.
- Canut, H.; Albenne, C.; Jamet, E. Post-translational modifications of plant cell wall proteins and peptides: A survey from a proteomics point of view. Biochim. Biophys. Acta 2016, 1864, 983–990.
- Duruflé, H.; Hervé, V.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Proline hydroxylation in cell wall proteins: Is it yet possible to define rules? Front. Plant Sci. 2017, 8, 1802.
- Tan, L.; Varnai, P.; Lamport, D.T.; Yuan, C.; Xu, J.; Qiu, F.; Kieliszewski, M.J. Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 2010, 285, 24575–24583.
- Paulick, M.; Bertozzi, C. The glycosylphosphatidylinositol anchor: A complex membrane-anchoring structure for proteins. Biochemistry 2014, 47, 6991–7000.
- Faye, L.; Boulaflous, A.; Benchabane, M.; Gomord, V.; Michaud, D. Protein modifications in the plant secretory pathway: Current status and practical implications in molecular pharming. Vaccine 2005, 23, 1770–1778.
- Gomord, V.; Fitchette, A.; Menu-Bouaouiche, L.; Saint-Jore-Dupas, C.; Plason, C.; Michaud, D.; Faye, L. Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 2010, 8, 564–587.
- Toustou, C.; Walet-Balieu, M.; Kiefer-Meyer, M.; Houdou, M.; Lerouge, P.; Foulquier, F.; Bardor, M. Towards understanding the extensive diversity of protein N-glycan structures in eucaryotes. Biol. Rev. 2022, 97, 732–748.
- Schoberer, J.; Strasser, R. Plant glyco-biology. Semin. Cell Dev. Biol. 2018, 80, 133–141.
- Strasser, R. Plant protein glycosylation. Glycobiology 2016, 26, 926–939.
- Strasser, R.; Seifert, G.; Doblin, M.; Johnson, K.; Ruprecht, C.; Pfrengle, F.; Bacic, A.; Estevez, J. Cracking the “sugar code”: A snapshot of N- and O-glycosylation pathways and functions in plants cells. Front. Plant Sci. 2021, 12, 640919.
- Mócsai, R.; Figl, R.; Troschl, C.; Strasser, R.; Svehla, E.; Windwarder, M.; Thader, A.; Altmann, F. N-glycans of the microalga Chlorella vulgaris are of the oligomannosidic type but highly methylated. Sci. Rep. 2019, 9, 33.
- Mathieu-Rivet, E.; Scholtz, M.; Arias, C.; Dardelle, F.; Schulze, S.; Le Mauff, F.; Teo, G.; Hochmal, A.; Blanco-Rivero, A.; Loutenier-Bourhiss, C.; et al. Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol. Cell. Proteom. 2013, 12, 3160–3183.
- Ruiz-May, E.; Sørensen, I.; Fei, Z.; Zhang, S.; Domozych, D.; Rose, J. The secretome and N-glycosylation profiles of the charophycean green alga, Penium margaritaceum, resemble those of embryophytes. Proteomes 2018, 6, 14.
- Remco, V.; Loutelier-Bourhis, C.; Fitchette, A.; Margerie, P.; Gonneau, M.; Faye, L.; Lerouge, P. Protein N-glycosylation is similar in the moss Physcomitrella patens and in higher plants. Planta 2003, 218, 269–275.
- Pattison, R.; Amtmann, A. N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 2009, 14, 92–99.
- Fanata, W.; Lee, K.; Son, B.; Yoo, J.; Harmoko, R.; Ko, K.; Ramasamy, N.; Kim, K.; Oh, D.; Jung, H.; et al. N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J. 2013, 73, 966–979.
- Strasser, R.; Altmann, F.; Mach, L.; Glössel, J.; Steinkellner, H. Generation of Arabidopsis thaliana plants with complex N-glycans lacking β1,2-linked xylose and core α1,3-linked fucose. FEBS Lett. 2004, 561, 132–136.
- Kolkas, H.; Balliau, T.; Chourré, J.; Zivy, M.; Canut, H.; Jamet, E. The cell wall proteome of Marchantia polymorpha reveals specificities compared to those of flowering plants. Front. Plant Sci. 2022, 12, 765846.
- Bu, T.; Shen, J.; Chao, Q.; Shen, Z.; Yan, Z.; Zheng, H.; Wang, B. Dynamic N-glycoproteome analysis of maize seedling leaves during de-etiolation using Concanavalin A lectin affinity chromatography and a nano LC-MS/MS-based iTRAQ approach. Plant Cell Rep. 2017, 36, 1943–1958.
- Chang, Y.; Zhu, D.; Duan, W.; Deng, X.W.; Zhang, J.; Ye, X.; Yan, Y. Plasma membrane N-glycoproteome analysis of wheat seedling leaves under drought stress. Int. J. Biol. Macromol. 2021, 193, 1541–1550.
- Zhang, M.; Chen, G.; Lv, D.; Li, X.; Yan, Y. N-linked glycoproteome profiling of seedling leaf in Brachypodium distachyon L. J. Proteome Res. 2015, 14, 1727–1738.
- Geng, F.; Liu, X.; Wang, J.; He, R.; Zhao, J.; Xiang, D.; Zou, L.; Peng, L.; Zhao, G. In-depth mapping of the seed phosphoproteome and N-glycoproteome of Tartary buckwheat (Fagopyrum tataricum) usinf off-line pH RPLC fractionation and nLC-MS/MS. Int. J. Biol. Macromol. 2019, 137, 688–696.
- Ruiz-May, E.; Hucko, S.; Howe, K.; Zhang, S.; Sherwood, R.; Thannhauser, T.; Rose, J. A comparative study of lectin affinity based plant N-glycoproteome profiling using tomato fruit as a model. Mol. Cell. Proteom. 2014, 13, 566–579.
- Zhang, X.; Tang, H.; Du, H.; Liu, Z.; Bao, Z.; Shi, Q. Comparative N-glycoproteome analysis provides novel insight into the regulation mechanism in tomato (Solanum lycopersicum L.) during fruit ripening process. Plant Sci. 2020, 293, 110413.
- Catalá, C.; Howe, K.; Hucko, S.; Rose, J.; Thannhauser, T. Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using Concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics 2011, 11, 1530–1544.
- Kumar, S.; Kumar, K.; Pandey, P.; Rajamani, V.; Padmalatha, K.; Dhandapani, G.; Kanakachari, M.; Leelavathi, S.; Kumar, P.; Reddy, V. Glycoproteome of elongating cotton fiber cells. Mol. Cell. Proteom. 2013, 12, 3777–3789.
- Liu, Y.; Ma, L.; Cao, D.; Gong, Z.; Fan, J.; Hu, H.; Jin, X. Investigation of cell wall proteins of C. sinensis leaves by combining cell wall proteomics and N-glycoproteomics. BMC Plant Biol. 2021, 21, 384.
- Minic, Z.; Jamet, E.; Negroni, L.; der Garabedian, P.A.; Zivy, M.; Jouanin, L. A sub-proteome of Arabidopsis thaliana trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 2007, 58, 2503–2512.
- Zhang, Y.; Giboulot, A.; Zivy, M.; Valot, B.; Jamet, E.; Albenne, C. Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 2011, 72, 1109–1123.
- Song, W.; Mentik, R.; Henquet, M.; Cordewener, J.; van Dijk, A.; Bosch, D.; America, A.; van der Krol, A. N-glycan occupancy of Arabidopsis N-glycoproteins. J. Proteom. 2013, 93, 343–355.
- Zeng, W.; Ford, K.; Bacic, A.; Heazlewood, J. N-linked glycan micro-heterogeneity in glycoproteins of Arabidopsis. Mol. Cell. Proteom. 2018, 17, 413–421.
- Sultana, N.; Florance, H.; Johns, A.; Smirnoff, N. Ascorbate deficiency influences the leaf cell wall glycoproteome in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 375–384.
- Xu, S.; Medzihradszky, K.; Wang, Z.; Burlingame, A.; Chalkley, R. N-Glycopeptide profiling in Arabidopsis inflorescence. Mol. Cell. Proteom. 2016, 15, 2048–2054.
- Ligat, L.; Lauber, E.; Albenne, C.; San Clemente, H.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 2011, 11, 1798–1813.
- Ma, J.; Wang, D.; She, J.; Li, J.; Zhu, J.; She, Y. Endoplasmic reticulum-associated N-glycan degradation of cold-upregulated glycoproteins in response to chilling stress in Arabidopsis. New Phytol. 2016, 212, 282–296.
- Yoo, J.; Ko, K.; Seo, H.; Park, S.; Fanata, W.; Harmoko, R.; Ramasamy, N.; Thulasinathan, T.; Mengiste, T.; Lim, J.; et al. Limited addition of the 6-arm β1,2-linked N-acetylglucosamine (GlcNAc) residue facilitates the formation of the largest N-glycan in plants. J. Biol. Chem. 2015, 290, 16560–16572.
- Pedersen, C.; Loke, I.; Lorentzen, A.; Wolf, S.; Kamble, M.; Kristensen, S.; Munch, D.; Radutoiu, S.; Spillner, E.; Roepstorff, P.; et al. N-glycan maturation mutants in Lotus japonicus for basic and applied glycoprotein research. Plant J. 2017, 91, 394–407.
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8, 94.
- Duruflé, H.; Hervé, V.; Ranocha, P.; Balliau, T.; Zivy, M.; Chourré, J.; San Clemente, H.; Burlat, V.; Albenne, C.; Déjean, S.; et al. Cell wall adaptation of two contrasted ecotypes of Arabidopsis thaliana, Col and Sha, to sub-optimal growth conditions: An integrative study. Plant Sci. 2017, 263, 183–193.
- Shigeyama, T.; Watanabe, A.; Tokuchi, K.; Toh, S.; Sakurai, N.; Shibuya, N.; Kawakami, N. α-Xylosidase plays essential roles in xyloglucan remodelling, maintenance of cell wall integrity, and seed germination in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 5615–5629.
- Lannoo, N.; Van Damme, E. Review/N-glycans: The making of a varied toolbox. Plant Sci. 2015, 239, 67–83.
- Strasser, R.; Bondili, J.; Schoberer, J.; Svoboda, B.; Liebminger, E.; Glössl, J.; Altmann, F.; Steinkellner, H.; Mach, L. Enzymatic properties and subcellular localization of Arabidopsis β-N-acetylhexosaminidases. Plant Physiol. 2007, 145, 5–16.
- Dupoiron, S.; Zischeck, C.; Ligat, L.; Carbonne, J.; Boulanger, A.; de Bernonville, T.D.; Lautier, M.; Rival, P.; Jamet, E.; Lauber, E.; et al. The N-glycan cluster from Xanthomonas campestris pv. campestris: A toolbox for sequential plant N-glycan processing. J. Biol. Chem. 2015, 290, 6022–6036.
- Renzi, F.; Manfredi, P.; Mally, M.; Moes, S.; Jenö, P.; Cornelis, G. The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLoS Pathog. 2011, 7, 1002118.
- Brewer, C.; Bhattacharya, L. Concanavalin A interactions with asparagine-linked glycopeptides. The mechanisms of binding of oligomannose, bisected hybrid, and complex type carbohydrates. Glycoconj. J. 1988, 5, 159–173.
- Kasturi, L.; Eshleman, J.; Wunner, W.; Shakin-Eshleman, S. The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J. Biol. Chem. 1995, 270, 14756–14761.
- Mellquist, J.; Kasturi, L.; Spitalnik, S.; Shakin-Eshleman, S. The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 1998, 37, 6833–6837.
- Shakin-Eshleman, S.; Spitalnik, S.; Kasturi, L. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J. Biol. Chem. 1996, 271, 6363–6366.
More
