This entreviewy discusses recent advances in the development of biosensors for the purposes of malaria diagnostics. It underscore relevant challenges that have defined the gap between biosensor development and their successful utilization in routine clinical practice within resource-limitted settings. It proposes a way to think about developing biosensors that are suitable for biomedical diagnostics applications.
- Malaria Diagnostics
- Biosensors
- Point-of-Care Testing
- Infectious Diseases
- Hemozoin
- Malaria biomarkers
- Aldolase
- Glutamate Dehydrogenase
- Plasmodiumlactate dehydrogenas
- Histidine-rich protein 2
Biosensors and immunosensors have experienced unprecedented growth in recent years and seem to be the most promising sensing tools with several analytical benefits and cost efficiency [[1][2]]. This growth has been driven in part by the surge in demand for POC devices in clinical diagnosis where biological sensing is integrated with microelectronics to form portable analytical devices. To date, nearly sixty years after the first biosensor for glucose detection, the technology has been widespread in several fields of analyte detection [[3]]. Glucometers have evolved enormously, receiving vast commercial success [[4]] whereas biosensors for other diseases have been limited to experimental research. Among the types of sensors, electrochemical biosensors have received considerable interest in clinical diagnostics owing to key advantages in their design, assay simplicity, and superior analytical performance over conventional laboratory methods [[5][6]]. These qualities make them suitable for POC application amidst efforts to improve and miniaturize electrochemical systems for portable devices.
Most attempts to create miniaturized electrochemical devices for on-site analysis have applied screen-printed electrodes (SPE) as transducers and various nanomaterials as signal amplification strategies to improve the assay sensitivity [[7][8][9][10]]. Electrochemical immunosensors have been commonly applied to malaria diagnostic research given the benefits of low detection limits, wide linear response range, stability and reproducibility [[11]]. The strategies for detection comprise either a labelled assay in which apply amperometry and colorimetry or an impedimetric strategy, attractive for highly sensitive label-free detection [[12][13]]. Only a few potentiometric techniques have been reported [14]]. The performances of selected biosensors reported for malaria biomarkers detection is summarized in Table 1. The choice of PfHRP-2 and LDH is still predominant, similar to RDTs. However, there is an increase in preference for pLDH possibly due to the persistence of PfHRP-2 antigenemia for several weeks after parasite clearance [[15]] and reports of mutant strains from Africa and Asia with deleted PfHRP-2 genes [[16][17][18]].
Table 1. Summary of selected biosensors reporting detection of various malaria biomarkers.
Analytes | Sensing Technique/Response | Transducer | Biomarker | Receptor Molecule | LoD | Range | Response Time | Storage Stability | References | ||||||||||||||||||||||
Antigens | Colorimetric | - | pLDH (PvLDH, PfLDH) | pL1 aptamer | 8.3–8.7 pM (PvLDH) | 10.3–12.5 pM (PfLDH) | NA | NA | NA | [[19]] | |||||||||||||||||||||
EIS | Gold electrode | pLDH | pL1 aptamer | ** 108.5 fM for PvLDH | ** 120.1 fM for PfLDH | NA | NA | NA | [[20]] | ||||||||||||||||||||||
EIS | GCE | pLDH | P38 aptamer (90 mer ssDNA) | 0.5 fM | NA | - | NA | [[21]] | |||||||||||||||||||||||
EIS | GCE | HRP-2 | Anti-HRP-2 antibody | ** 6.8 ag/mL. | 10 ag/mL–10 mg/mL | NA | 2 months (86.5%) | [[22]] | |||||||||||||||||||||||
Chemiresistive (electrical conductance) | - | PfHRP-2 | Anti-HRP-2 antibody | 0.97 fg/mL | 10 fg/mL–10 ng/mL | NA | 15 days (94.2%) | [[9]] | |||||||||||||||||||||||
- | - | PfHRP-2 | Anti-PfHRP-2 | 0.025 ng/mL | 0.01–10 ng/mL | - | - | [[23]] | |||||||||||||||||||||||
EIS | Gold disc electrodes | PfGDH | ssDNA aptamer (NG3) | * 0.77 pM | 100 fM–100 nM | NA | NA | [[24]] | |||||||||||||||||||||||
Potentiometric (FET) | Gold micro-electrodes | PfGDH | ssDNA aptamer (NG3) | ** 16.7 pM | * 48.6 pM | 100 fM–10 nM | 5 s |
| [[14]] | ||||||||||||||||||||||
Amperometric | Gold-SPE | PfHRP-2 | Anti-PfHRP 2 mAb | ** 36 pg/mL | * 40 pg/mL | NA | NA | NA | [[25]] | ||||||||||||||||||||||
Amperometric | Gold-SPE | pLDH | pLDH capture antibody | ** 19 pg/mL | * 23 pg/mL | - | - | - | [[26]] | ||||||||||||||||||||||
Spectrophotometric | Indicator displacement medium | - | PfHRP-2 | NA | 30 ± 9.6 nM | 10–100 nM | 5 min | NA | [[27]] | ||||||||||||||||||||||
Colorimetric | - | PfLDH | 2008s-biotin DNA aptamer | ** 4.9 ng/mL | NA | <1h | 2 months | [[28]] | |||||||||||||||||||||||
Colorimetric | - | PfLDH | 2008s aptamer | - | NA | 20 min | - | [[29]] | |||||||||||||||||||||||
Amperometric | SPE | PfHRP-2 | Mouse anti-PfHRP-2 antibody | ** 8 ng/mL | NA | NA | NA | [[30]] | |||||||||||||||||||||||
FRET | - | pLDH | Fluorescently-labeled aptamer (36 mer ssDNA) | ** 550 pM | NA | NA | NA | [[31]] | |||||||||||||||||||||||
Amperometric magneto Immunosensor | - | PfHRP2 | Anti-HRP2 IgM Antibody | 0.36 ng/mL | 0.35–7.8 ng/mL | NA | NA | [[32]] | |||||||||||||||||||||||
Antibodies | SPR | Gold disc | Antibodies of Pf. | PfHRP2 | ** 5.6 pg for mAb | - | NA | NA | [[33]] | ||||||||||||||||||||||
Nucleic acids | Quartz Crystal Microbalance | - | Pf msp2 gene | Biotinylated probe | ≥0.025 ng/mL of target DNA | NA | NA | 180 days | [[34]] | ||||||||||||||||||||||
Droplet Microfluidic Platform | - | Pf topoisomerase I | ds DNA substrate | NA | NA | NA | NA | [[35]] | |||||||||||||||||||||||
SERS Nanoplatform | - | Pf DNA sequences | Magnetic bead and nanorattle | 100 attomoles | 10−11–10−10 M | NA | NA | [[36]] | |||||||||||||||||||||||
Quartz Crystal Microbalance | Silver electrode | 18s rRNA gene (Pf and Pv) | immobilized probe | - | - |
|
| [[37]] | |||||||||||||||||||||||
Infected red blood cells | EIS | SPE | Pf infected RBCs | monoclonal antibody | - | 102–107 cells/mL | NA | NA | [[38]] | ||||||||||||||||||||||
microfluidic separation and MRR | - | Infected RBCs | - | 0.0005% parasitemia | - | - | - | [[39]] |
LoD: limit of detection; * LoD: LoD in real samples; ** LoD: LoD in buffer; EIS: electrochemical impedance spectroscopy; FET: field effect transistor; FRET: fluorescence resonance energy transfer; GCE: glassy carbon electrode; MRR: magnetic resonance relaxometry; SPE: screen-printed electrode; SERS: surface-enhanced Raman spectroscopy; SPR: surface plasmon resonance; SWV: square wave voltammetry.
Detection of PfHRP-2 in Clinical Samples
Histidine-rich protein 2 (HRP-2) is specific to P. falciparum (PfHRP-2) and is secreted into peripheral blood during parasite growth and development where it plays a role in heme detoxification. The antigen’s widespread application in electrochemical and optical immunosensors is due to copious expression levels throughout the parasite life cycle. Although primarily abundant in blood, trace amounts can be found in cerebrospinal fluid, urine, and saliva of infected patients [[40][41]], which offer an opportunity for non-invasive testing. Nonetheless, blood is preferred because of small sample volumes required to target the antigen. Painless testing has attractive public health benefits of voluntary testing and participation in screening programs geared towards malaria control [[42]]. However, only a few publications have attempted urine or saliva analysis as noninvasive malaria diagnostics [[43]].
Electrochemical techniques have been shown to outperform optical methods in many modelled POC tests. Nanoparticles, primarily gold (AuNP) have been adopted in signal amplification for amperometric immunosensors [[44][45][46]]. Their small size and ease of immobilizing bioconjugate probes allow for increased surface concentration of enzyme-tagged detection antibodies, hence higher signals from the catalytic reaction of enzyme and substrate. Sharma et al. were first to report an electrochemical immunosensor to detect PfHRP-2 in blood by amperometry [65]. The disposable immunosensor utilized multi-walled carbon nanotubes (MWCNTs) and gold nanoparticles (Nano-Au) to modify screen printed electrodes (SPE); resulting in Nano-Au/MWCNT/SPEs onto which rabbit-derived anti-PfHRP-2 were immobilized as capture antibodies. A sandwich enzyme-linked immunosorbent assay format was employed for the biosensor with alkaline phosphatase (ALP)-conjugated antibodies. Amperometric measurements were applied using ALP hydrolysis of 1-naphthyl phosphate. The Nano-Au/MWCNT/SPE had a limit of detection (LoD) of 8.0 ng/mL (compared to 80.0 ng/mL for bare SPE and 20.0 ng/mL for MWCNT/SPE) (Table 1). This enhanced performance was attributable to the synergistic effect of MWCNTs and AuNP. More importantly, the immunosensor had a superior analytical performance compared with a commercial immunochromatographic lateral flow test in the analysis of microscopy positive patient sample (sensitivity: 96% vs. 79%, specificity; 94% vs. 81% respectively).
In assessing exposure to malaria parasites, recombinant PfHRP-2 was used as a recognition element for anti-PfHRP-2 antibodies in an amperometric immunosensor for early stages of malaria and at low parasitemia [[47]]. SPEs were modified with alumina sol-gel (Al2O3) and AuNP to obtain AuNP/Al2O3 sol-gel/SPE after which PfHRP-2 was bound. Rabbit anti-PfHRP-2 and anti-rabbit IgG-ALP conjugate were directed against capture antigens and the analytical responses determined by amperometry. In comparison to ‘gold standard’ microscopy, the immunosensor exhibited a sensitivity of 92% and a specificity of 90%. In another study that detected monoclonal antibodies to recombinant PfHRP-2 (MoaPfHRP-2), a gold chip was pre-treated with 4-mercaptobenzoic to immobilize recombinant PfHRP-2, then monitored for interactions between the antigen and antibody [68]. Label free surface plasmon resonance (SPR) screening of the interaction between the recombinant protein and target antibody produced a LoD of 5.6 pg (Table 1).
Magnetic nanoparticles (MNPs) have also been applied in the development of a highly sensitive malaria immunosensor. Anti-HRP-2 was covalently attached to MNPs as capture elements and a second monoclonal antibody that binds a different epitope of the target antigen was labelled with horse radish peroxidase (HRP) to provide an electrochemical signal [67]. The anti-HRP-2-MNPs were captured onto a magnetic graphite-epoxy composite electrode incubated with HRP-2-spiked serum and anti-HRP-2-HRP in a sandwich assay format. Amperometric measurements produced an LoD of 0.36 ng/mL (Table 1), much lower than Sharma et al. [65] reported. Translating this strategy to the field would require magnetic supports for electrodes [67].
More recently, Hemben et al. used anti-PfHRP-2 monoclonal antibodies to capture PfHRP-2 at the surface of a screen printed gold electrode (Table 1) [60]. The captured antigen was targeted with HRP-labelled antibodies and the quantification of PfHRP-2 derived from the substrate (TMB-H2O2)-enzyme reaction by amperometry (Figure 2A). The LoDs in buffer and spiked human samples were determined as 2.14 ng/mL and 2.95 ng/mL, respectively. Labelled antibodies were subsequently conjugated to gold nanoparticles (AuNP) to amplify the sensor signal which improved the sensitivity and LoD in buffer (36.0 pg/mL) and spiked serum samples (40.0 pg/mL).
While biosensing platforms for most disease biomarkers tend to rely on antibodies as capture molecules, a challenge with immunoassays is related to antibody stability, a prerequisite. Some attempts to circumvent these drawbacks have included genetic manipulations that improve the stability and shelf-life of antibodies and the use of synthetic alternatives such as aptamers. For example, researchers cloned and expressed cDNA fragments encoding the variable domains (VL-CL and VH-CH1) of two monoclonal antibodies against PfHRP-2 (F1546 and F1110) in Escherichia coli [[47]]. The recombinant Fab fragments showed similar binding properties to those of the parental monoclonal antibodies (mAb). This approach proposes a cost-efficient alternative to large-scale antibody production for diagnostic application with the opportunity of engineered antibody fragments with improved affinity, stability, and resistance to denaturation even with prolonged storage in uncontrolled temperatures in the field.
Besides using molecular labels and nanoparticles for improved diagnostics, some assays can probe the intimate recognition between the receptor and target alone. The benefits of using such label-free formats include a reduction in the assay complexity, preparation time, and analysis cost as it eliminates potentially confounding chemical labels. These strategies are better suited to field applications and under resourced settings where laboratories and skilled personnel are unavailable. A label-free a piezoelectric immunosensor for PfHRP-2 was designed by. applying mixed self-assembled monolayers (SAMs) of thioctic acid and 1-dodecanethiol on gold quartz crystal microbalance (QCM) Anti-PfHRP-2 antibodies were covalently immobilized unto the SAM-modified electrodes via EDC-NHS activation and the frequency change resulting from binding of different concentrations of PfHRP-2 measured [[48]]. The immunosensor exhibited a LoD of of 12.0 ng/mL with a linear range of 15.0–60.0 ng/mL for analysis of PfHRP-2 in buffer. This was higher than their previously reported amperometric immunosensor (8 ng/mL [[30]] vs. 12ng/mL [[48]]), and weaker responses were observed for PfHRP-2 concentrations lower than 25.0 ng/mL. However, application of the sensor to clinical samples produced comparable sensitivities with a commercial immunochromatographic test (ICT) kit (NOW® Malaria, Binax, Inc., Scarborough, Maine, USA).
An indicator displacement assay (IDA) was used to detect and quantify HRP-2 in sera [[27]]. The label-free spectrophotometric method did not require any biorecognition elements and was based on the color change of murexide either in complex with nickel (Ni2+) or free in solution. In the IDA, competition between HRP-2 and Ni2+ displaces murexide from the murexide-Ni2+ complex. The resultant color intensity was proportional to free unbound murexide is measured to quantify HRP-2. The assay had LoD of 30.0 ± 9.6 nM (dynamic range of 10–100.0 nM) without any interfering signals from serum proteins (Table 1).
Electrochemical impedance spectroscopy (EIS)-based methods present with numerous advantages that make them suitable candidates for POC application [[49]]. Their high sensitivity is evident from lower LoDs they tend to produce compared with other electrochemical methods. The lowest LoD yet reported for malaria was achieved by impedimetric detection of PfHRP-2 [[22]] (Table 1). In the sensor design, copper doped zinc oxide nanofibers (CZnONF) was drop-casted on glassy carbon electrode (GCE/CZnONF) followed by SAM modification and chemisorption of anti-PfHRP-2 (Figure 1B). The highly sensitive nanosensor (28.5 kΩ/(g/mL)/cm3) attained a detection limit of 6.8 ag/mL. The authors subsequently reported a flexible chemiresistive immunosensor in which the transducer comprised a 1-dimensional MWCNT-zinc oxide (MWCNTs-ZnO) nanofiber drop-casted on micro gold electrodes [[9]] (Table 1). Capture antibodies, anti-PfHRP-2 were immobilized on MWCNTs-ZnO by EDC-NHS crosslinking and resistance changes (ΔR) measured to monitor the formation of PfHRP-2-anti-PfHRP-2 complexes. The device demonstrated good analytical performance as well as potential towards the development of POCTs with a linear response ranging from 10 fg/mL to10 ng/mL, LoD of 0.97 fg/mL and high specificity for PfHRP-2 over non-specific antigens.
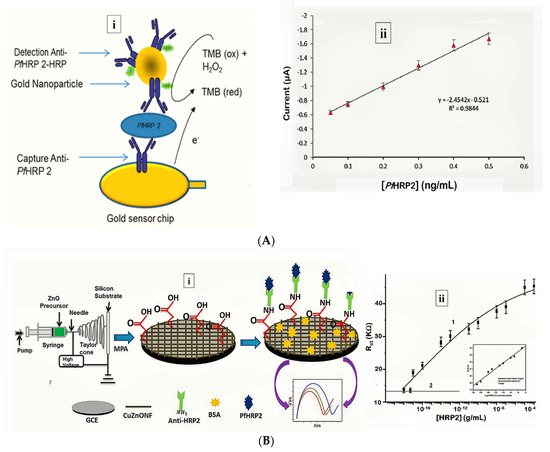
Figure 1. Strategy for (A) labelled amperometric [60] and (B) label free impedimetric [[22]] electrochemical detection of PfHRP-2. (A) (i) Gold nanoparticle amplified sandwich assay and (ii) plot of chronoamperometric response of PfHRP-2 detection in spiked serum (0.05–0.5 ng/mL PfHRP). Reprinted from Hemben et al. [[25]] with permission from MDPI. (B) (i) CZnONF dispersions drop-casted onto GCE followed by chemisoption of anti-HRP2 unto MPA treated electrodes and (ii) calibration curve of impedimetric responses obtained after incubating GCE/fCuZnONFs/Anti-HRP2 biosensor with varying concentrations of PfHRP2 (10 ag/mL–10 µg/mL). (Adapted from Paul et al. [57] with permission from Elsevier).
Detection of pLDH in Clinical Samples
Plasmodium lactate dehydrogenase (pLDH) plays a catalytic role in the glycolytic pathway during the intraerythrocytic stages of Plasmodium. It is produced by metabolically active parasites within infected red blood cells (RBCs) [[50][51][52]] and has conserved catalytic residues in all Plasmodium spp. except in P. knowlesi. Unlike HRP-2, pLDH is indicative of a recent infection and is generally cleared within 24 h of parasite clearance; hence, it more reliable in identifying recent unresolved infections.
There seems to be a growing trend of aptamer-based sensors targeting pLDH [[19],[20],[53]]. Compared to antibodies, aptamers are smaller in size, thermostable, extended shelf life without functional degradation, affordable, easily synthesized and can be readily modified. It could, to an extent be an alternative remedy in overcoming difficulties associated with using antibody-based tests. The unit costs for malaria RDTs in many African countries falls within the unsubsidized range of USD 2.54–2.83. Yet, a study in Uganda revealed that consumers were willing to pay an average of USD 0.53 [[54]]. The prospects of some biosensors/aptasensors being estimated at 1 USD or less per test, proposes a significant reduction to the general healthcare costs in impoverished tropical regions where malaria is prevalent.
In a prospective tool for asymptomatic and early diagnosis of malaria, single-stranded DNA aptamers (pL1 aptamers) were used to target recombinant Plasmodium falciparum LDH (PfLDH) and Plasmodium vivax LDH (PvLDH) in buffer and in real samples [55] (Table 1). Impedance measurements for the interaction between pL1 and the target proteins demonstrated high sensitivity and specificity with LoDs measuring 108.5 fM (for PvLDH) and 120.1 fM (PfLDH). Native pLDH in clinical samples was also detected up to 1 parasite/µL.
Figueroa-Miranda et al. immobilized 2008s aptamers on a SAM-modified gold electrode to bind pLDH down to the detection limit of 0.84 pM in buffer and 1.30 pM in blood (Figure 2, Table 1) [[53]]. The aptasensor had a detection range between 1 pM–10 nM and remained highly selective for PfLDH even in the presence of high concentrations of serum proteins and analogues of LDH from muse muscle. The thiol-gold covalent bonding from SAMs conferred high immobilization stability to the aptamer and could be regenerated and re-used for up to three times without loss of analytical performance. A major observation was the influence of isoelectric point (pI) of PfLDH on impedimetric responses which tended to increase where pH > pI and a decrease at pH < pI. A likely reason for this occurrence is attributable to a repulsion and attraction of the redox probe to the electrode surface.
Aptamer functionalized microbeads were used to determine the capture and measure the intrinsic enzymatic activity of LDH in a colorimetric assay (Figure 3A) [[27]]. In the aptamer-tethered enzyme capture (APTEC) assay, the beads conferred a wide surface area for analyte binding to produce a LoD of 4.9 ng/mL for recombinant PfLDH (Table 1). Further work integrated the APTEC assay into a portable microfluidic biosensor (Figure 3B) [[29]]. The platform resolved some of the assay’s initial limitations of large sample and reagent volumes while detecting P. falciparum with high specificity and sensitivity in cultures and clinical samples.
Fluorescently-labelled aptamers were adsorbed to molybdium disulphide (MoS2) nanosheets to develop a FRET aptasensor which selectively detected pLDH in a heterogeneous protein mixture [[31]]. The mechanism of the assay was based on a “capture-release” model whereby fluorescence of the aptamer is quenched upon aptamer-MoS2 nanosheets binding and restored in the presence of pLDH when the aptamer is released from the nanosheets. The attachment and detachment processes are facilitated by the high affinity between aptamers and pLDH. The sensor achieved LoD of 550.0 pM (Table 1).
Hemben et al. functionalized screen-printed gold electrodes (SPGE) with anti-pLDH antibodies and applied a sandwich assay format to detect pLDH [61]. The sensor initially achieved LoDs of 1.80 ng/mL in buffer and 0.70 ng/mL in serum. Application of colloidal AuNPs functionalized with HRP-labelled detection antibodies enhanced amperometric signals to LoDs down to 19 pg/mL (in buffer) and 23 pg/mL (in serum) (Table 1).
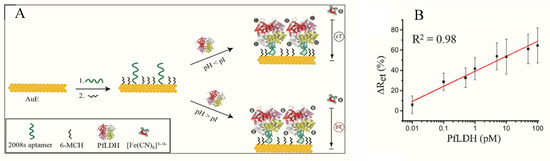
Figure 2. (A) Schematic representation of the PfLDH aptasensor and (B) calibration plot for 0.01 pM–10 nM PfLDH in 5 mM [Fe(CN)6]3−/4− solution at pH 7.5. (Reprinted from Figueroa-Miranda et al. [[54]] with permission from Elsevier).
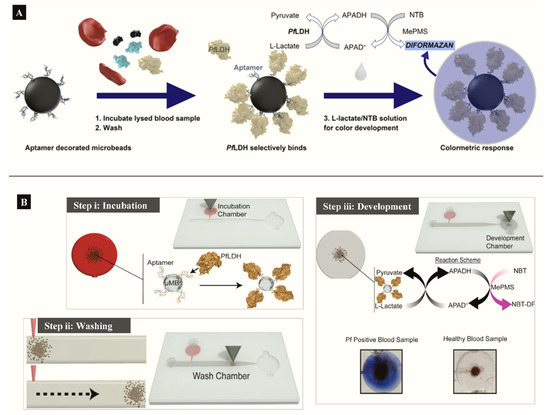
Figure 3. (A) APTEC biosensor based on capturing PfLDH and using its enzymatic activity to produce a colorimetric assay (Reprinted from Dirkzwager et al. [63] with permission from the American Chemical Society). (B) Working principle ((i) incubation, (ii) washing (ii) and (iii) color development) of a simple portable 3D-printed microfluidic device for diagnosis of malaria in clinical samples constructed from the APTEC assay (Reproduced from Fraser et al. [64] with permission from Elsevier).
Detection of GDH in Clinical Samples
Ubiquitous enzymes such as glutamate dehydrogenases (GDH) in Plasmodium parasites play a role in glutamate catabolism and ammonium assimilation [[55],[56][57][58]]. The enzyme is present throughout the sexual and asexual stages of the parasite development in significantly soluble quantities [[59]]. Several structural and kinetic distinctions between host and parasite GDH makes it potentially useful in targeting live parasites [[60]].
A label-free capacitive aptasensor was constructed by graftting thiolated ssDNA aptamer (NG3) specific to P. falciparum (PfGDH) on a gold electrode [[24]]. The sensor produced a LoD of 0.77 pM in serum with dynamic range 100 fM−100 nM (Table 1). Subsequently, the authors integrated the process into an extended gate field effect transistor (EgFET).The NG3 aptamers were immobilized on an inter-digitated gold microelectrodes (IDµE) and connected to the FET to construct a sensitive and stable miniaturized aptaFET biosensor (Figure 4) [[14]]. A benefit of FET-type systems is that, it enables for sensitive and simple electrochemical measurements without requiring a typical redox marker [[61]]. Following calibration curves in varying concentrations of PfGDH-spiked buffer and serum, a linear detection range of 100 fM–10 nM was obtained with LoDs in buffer and serum being 16.7 pM and 48.6 pM, respectively (Table 1). The FET-based potentiometric sensor was highly selective in the presence of analogous human and plasmodial proteins, making it suitable for analysis of real sample for malaria diagnosis.
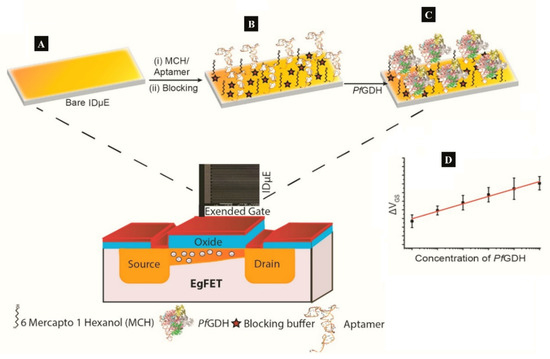
Figure 4. (A–C) Strategy for the label free aptaFET and (D) calibration curves obtained for PfGDH detection in serum. (Adapted from Singh et al. [[14]] with permission from Elsevier).
Detection of Aldolase
Aldolase plays a key role in the glycolytic pathway of Plasmodium species where it catalyzes cleavage of fructose-1,6-bisphosphate into glyceraldehyde-3-phosphate and dihydroxyacetone phosphate [[62]]. Targeting aldolase as an antigens in malaria has been largely confined to ICTs, however, an evaluation of four aldolase- and LDH-based commercial ICTs found variations in specificity to P. vivax [[63]]. A probable reason why aldolase biosensors have not received much interest could be due to the poor sensitivity reported in aldolase ICTs. The genes encoding aldolase in P. falciparum and P. vivax are highly conserved [[64]], making it a poor marker of differential diagnosis. This adds on to the growing recommendations of paralleled detection of malaria antigens in test devices in order to maximize sensitivity and specificity while reducing the risk of misdiagnosis.
Detection of Hemozoin in Clinical Samples
At the erythrocytic stage of its life cycle, the malaria parasites digest about 60–80% of erythrocytic hemoglobin resulting in the formation of heme and polymerized to insoluble hemozoin crystallites [[65]]. Hemozoin is localized in parasite digestive vacuoles, therefore its presence in blood indicates a good marker of metabolically active Plasmodium parasites. The potential of surface-enhanced Raman spectroscopy (SERS) has been explored and shown to enhance the Raman signal of hemozoin by several folds [[66]]. Exposure of parasitized RBCs to a gold-coated butterfly wing as SERS substrate produced Raman shift within malarial hemozoin pigment whereas uninfected lysates did not. The spectral markers of hemozoin from infected RBC were detectable at the early-ring stage parasitemia levels of between 0.0005% and 0.005%. While enhancements of Raman signals occurs when hemozoin crystals are in direct contact with metal surfaces [[67]], another SERS method that applied synthesized silver nanoparticles inside parasites to achieve a close contact with hemozoin demonstrated an ultrasensitive hemozoin detection at 0.00005% parasitemia level in the ring stage (2.5 parasites/µL). These SERS methods have shown potential in early malaria diagnosis at low parasitemia levels, however, Raman spectrometers, and particularly those with high spectral resolutions, are expensive.
The paramagnetic properties of hemozoin crystals have been exploited for label-free detection using magnetic resonance relaxometry (MRR). In combination with a microfluidic setup, the MRR system achieved an accurate early detection at a parasitemia level of 0.0005% (Table 1) [[39]].
Detection of Other Relevant Malaria Biomarkers
Until date, most malaria biosensors have been principally confined to the established markers. Some other likely candidate targets have been speculated; heat-shock protein 70 (Hsp70), dihydrofolate reductase (DHFR)–thymidylate synthase (TS), heme-detoxification protein (HDP), glutamate-rich protein, hypoxanthine phosphoribosyl transferase, and phosphoglycerate mutase [[68][69][70]].
The capture of parasitized RBCs has been proposed as an alternative in overcoming the paucity of known malaria biomarkers. Even at low parasitemia, the populations of parasitized RBCs are elevated, reaching about 10,000 cells/µL in 0.2% parasitemia and 250,000–500,000 infected cell/mL in 5%–10% parasitemia [[71]]. Based on this knowledge, a novel microfluidic SELEX (I-SELEX) was implemented to identify a variant set of aptamers that distinctly bound different epitopes present on parasitized RBC surfaces [[72]]. In another study, researchers immobilized monoclonal antibodies on a AuNP modified screen-printed electrode as capture elements for malaria-infected cells [[12]] (Figure 5). Impedimetric changes caused by the interaction of monoclonal antibodies and parasitized RBCs distinguished infected from normal uninfected RBCs, measurable over a linear response concentration range of 102–108 cells/mL of infected RBCs (Table 1).
In another study, a highly sensitive and inexpensive biosensor was modelled to detect antibodies specific to P. vivax in serum (Table 1) [[73]]. Screen-printed carbon electrodes were coated with carbon nanotubes (CNTs) and EDC-NHS used to immobilize circumsporozoite protein (CSP) and thrombospondin related anonymous protein (TRAP). The assay detected anti-CSP and anti-P. vivax TRAP (anti-PvTRAP) antibodies directed against both antigens as low as 6–50 pg/L (concentrations in the range of 10–15 M) by means of EIS. Overall, the method demonstrated a promising approach to real-time probing of antibodies in serum directly without the need for sample preparation [[73]].
Nucleic acid markers have been exploited as reliable alternatives to proteins and antibodies in malaria diagnostics [[74]]. A DNA biosensor based on quartz crystal microbalance (QCM) was designed to targeted merozoite surface protein 2 (msp2) genes in P. falciparum [69]. The post-PCR label-free biosensor used avidin-biotin interactions to immobilize biotinylated probes complementary to the msp 2 gene unto gold QCM. Initial validation on laboratory strains showed probe-target binding at concentrations from 25–250 ng/mL and LoD of 0.025 ng/mL as well as genotyping potential (Table 1). Application of clinical samples confirmed no cross-reaction between species. In other studies, silver fabricated QCM platforms were applied to detect specific DNA fragments [[75]] and distinguish 18s rRNA gene of P. falciparum and P. vivax [72]. Both of these sensors were sensitive and could significantly differentiate species and mixed infections at the molecular level. Considering the hazards of UV visualizations and staining associated with agarose gel electrophoresis, the malaria gold/silver QCM promises a safer alternative. However, QCM methods still require prior amplification of genes which may not be feasible for POCT or immediate field uptake in under-resourced endemic areas where the capacity to perform PCR is lacking. A SERS-based method proposed by Ngo et al. to detect P. falciparum DNA fragments is suitable for automation and integration into portable molecular POCTs [[36]]. The assay comprised two complementary probes; a capture and reporter hybridized to magnetic beads and nanoratles (SERS tag) respectively, used to detect specific DNA in a sandwiched hybridization. The magnetic bead-DNA sequence-nanorattle formed in the presence of P. falciparum DNA target sequences is then concentrated for SERS measurement. The assay produced a LoD of 100 attomoles DNA (Table 1).
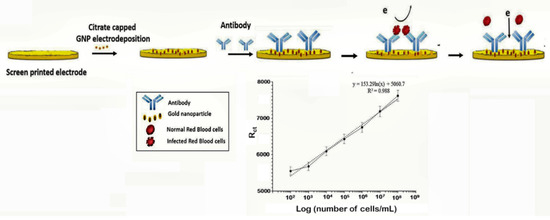
Figure 5. Schematic design of the impedimetric sensor malaria-infected RBCs and corresponding calibration plot. (Reprinted from Kumar et al. [[12]] with permission from the Royal Society of Chemistry).
Multi-Panel Biomarker Arrays for Malaria Detection
Simultaneous analysis of biomarkers maximizes the use of samples and gives a wide range of results that significantly improves test accuracy. Increased throughput, reduced reagents/assay setup and less labor are among features that could decrease assay errors and make parallel testing ideal for healthcare delivery. Moreover, the benefit directly translates to convenient sampling and low cost to providers and patients at the same time ensuring optimum outcome. A combination of antigens and genes have been reported in ICT and nucleic-acid-based methods respectively and are discussed elsewhere [[76][77][78]]. Multiplexed malaria testing is aimed primarily at differential diagnosis between parasite species. The most common methods combine PfHRP-2/LDH or PfHRP-2/aldolase to distinguish passive/resolved or active infections as well as differentiate falciparum from non-falciparum malaria [[79]]. Other multiplexed systems involving malaria antigens aim to differentiate malaria from other febrile illnesses (malaria/typhoid [[80]] and malaria/dengue [[81]]) since febrile presentations tend to mimic malaria clinical course leading to over-diagnosis and presumptive treatment of malaria.
References
- Muzyka, K. Biosensors and Bioelectronics Current trends in the development of the electrochemiluminescent immunosensors. Biosens. Bioelectron. 2014, 54, 393–407.
- Anthony Turner; Biosensors: sense and sensibility. Chemical Society Reviews 2013, 42, 3184, 10.1039/c3cs35528d.
- Mascini, M. A Brief Story of Biosensor Technology. In Biotechnological Applications of Photosynthetic Proteins: Biochips, Biosensors and Biodevices; Springer US: Boston, MA, USA, 2007; ISBN 978-0-387-33009-9.
- Thomas Ming Hung Lee; Over-the-Counter Biosensors: Past, Present, and Future. Sensors 2008, 8, 5535-5559, 10.3390/s8095535.
- María Soledad Belluzo; María Élida Ribone; Claudia Lagier; Assembling Amperometric Biosensors for Clinical Diagnostics. Sensors 2008, 8, 1366-1399, 10.3390/s8031366.
- Che-Wei Hsu; Fang-Ci Su; Po-Yu Peng; Hong-Tsu Young; Mike Yang; Gou-Jen Wang; A Novel Non-Enzymatic Electrochemical Glucose Biosensor Based on a Simple Lithographic Process. Volume 5B: 39th Mechanisms and Robotics Conference 2015, null, , 10.1115/detc2015-46954.
- Majid Kalate Bojdi; Mohammad Hossein Mashhadizadeh; Mohammad Behbahani; Ali Farahani; Saied Saeed Hosseini Davarani; Akbar Bagheri; Synthesis, characterization and application of novel lead imprinted polymer nanoparticles as a high selective electrochemical sensor for ultra-trace determination of lead ions in complex matrixes. Electrochimica Acta 2014, 136, 59-65, 10.1016/j.electacta.2014.05.095.
- Hadi Hosseini; Mohammad Behbahani; Mojtaba Mahyari; Hanif Kazerooni; Akbar Bagheri; Ahmad Shaabani; Ordered carbohydrate-derived porous carbons immobilized gold nanoparticles as a new electrode material for electrocatalytical oxidation and determination of nicotinamide adenine dinucleotide. Biosensors and Bioelectronics 2014, 59, 412-417, 10.1016/j.bios.2014.02.046.
- Brince Paul K; Asisa Kumar Panigrahi; Vikrant Singh; Shiv Govind Singh; A multi-walled carbon nanotube–zinc oxide nanofiber based flexible chemiresistive biosensor for malaria biomarker detection. The Analyst 2017, 142, 2128-2135, 10.1039/c7an00243b.
- Pratima R. Solanki; Ajeet Kaushik; Ved V. Agrawal; B. D. Malhotra; Nanostructured metal oxide-based biosensors. NPG Asia Materials 2011, 3, 17-24, 10.1038/asiamat.2010.137.
- Faridbod, F.; Gupta, V.K.; Zamani, H.A.; Electrochemical Sensors and Biosensors.. nt. J. Electrochem. 2011, , 2011, , 1–2..
- Binod Kumar; Vijayender Bhalla; Ravi Pratap Singh Bhadoriya; C Raman Suri; Grish C. Varshney; Label-free electrochemical detection of malaria-infected red blood cells. RSC Advances 2016, 6, 75862-75869, 10.1039/C6RA07665C.
- Clotilde Ribaut; Karine Reybier; Olivier Reynes; Jérôme Launay; Alexis Valentin; Paul Louis Fabre; Françoise Nepveu; Electrochemical impedance spectroscopy to study physiological changes affecting the red blood cell after invasion by malaria parasites. Biosensors and Bioelectronics 2009, 24, 2721-2725, 10.1016/j.bios.2008.12.018.
- Naveen K. Singh; Phurpa Dema Thungon; Pedro Estrela; Pranab Goswami; Phurpa Dema Thungoan; Development of an aptamer-based field effect transistor biosensor for quantitative detection of Plasmodium falciparum glutamate dehydrogenase in serum samples. Biosensors and Bioelectronics 2019, 123, 30-35, 10.1016/j.bios.2018.09.085.
- H Noedl; Chansuda Wongsrichanalai; Robert Scott Miller; Khin Saw Aye Myint; Sornchai Looareesuwan; Yaowalark Sukthana; Varee Wongchotigul; Herwig Kollaritsch; Gerhard Wiedermann; Walther H. Wernsdorfer; et al. Plasmodium falciparum: effect of anti-malarial drugs on the production and secretion characteristics of histidine-rich protein II.. Experimental Parasitology 2003, 102, 157-163, 10.1016/s0014-4894(03)00051-1.
- Ousmane A. Koita; Ogobara K. Doumbo; Amed Ouattara; Lalla K. Tall; Aoua Konaré; Mahamadou Diakité; Mouctar Diallo; Issaka Sagara; Godfred L. Masinde; Safiatou N. Doumbo; Amagana Dolo; Anatole Tounkara; Issa Traoré; Donald J. Krogstad; False-Negative Rapid Diagnostic Tests for Malaria and Deletion of the Histidine-Rich Repeat Region of the hrp2 Gene†. The American Journal of Tropical Medicine and Hygiene 2012, 86, 194-198, 10.4269/ajtmh.2012.10-0665.
- P. Serlemitsos; A Gas Proportional Chamber for use in Cosmic X-Ray Research. Symposium - International Astronomical Union 1971, 41, 213, 10.1017/s0074180900103560.
- Praveen Kumar Bharti; Himanshu Singh Chandel; Amreen Ahmad; Sri Krishna; Venkatachalam Udhayakumar; Mrigendra Pal Singh; Prevalence of pfhrp2 and/or pfhrp3 Gene Deletion in Plasmodium falciparum Population in Eight Highly Endemic States in India. PLOS ONE 2016, 11, e0157949, 10.1371/journal.pone.0157949.
- Weejeong Jeon; SeongHwan Lee; Manjunatha Dh; Changill Ban; D H Manjunatha; A colorimetric aptasensor for the diagnosis of malaria based on cationic polymers and gold nanoparticles. Analytical Biochemistry 2013, 439, 11-16, 10.1016/j.ab.2013.03.032.
- SeongHwan Lee; Kyung-Mi Song; Weejeong Jeon; Hunho Jo; Yoon-Bo Shim; Changill Ban; A highly sensitive aptasensor towards Plasmodium lactate dehydrogenase for the diagnosis of malaria. Biosensors and Bioelectronics 2012, 35, 291-296, 10.1016/j.bios.2012.03.003.
- Priyamvada Jain; Smita Das; Babina Chakma; Pranab Goswami; Aptamer-graphene oxide for highly sensitive dual electrochemical detection of Plasmodium lactate dehydrogenase. Analytical Biochemistry 2016, 514, 32-37, 10.1016/j.ab.2016.09.013.
- Brince Paul, K.; Kumar, S.; Tripathy, S.; Vanjari, S.R.K.; Singh, V.; Singh, S.G.; A highly sensitive self assembled monolayer modified copper doped zinc oxide nanofiber interface for detection of Plasmodium falciparum histidine-rich protein-2: Targeted towards rapid, early diagnosis of malaria. Biosens. Bioelectron. 2016, , 80, , 39–46..
- Emmanuel Gikunoo; Adeyabeba Abera; Eyassu Woldesenbet; A Novel Carbon Nanofibers Grown on Glass Microballoons Immunosensor: A Tool for Early Diagnosis of Malaria. Sensors 2014, 14, 14686-14699, 10.3390/s140814686.
- Naveen K. Singh; Sunil Arya; Pedro Estrela; Pranab Goswami; Capacitive malaria aptasensor using Plasmodium falciparum glutamate dehydrogenase as target antigen in undiluted human serum. Biosensors and Bioelectronics 2018, 117, 246-252, 10.1016/j.bios.2018.06.022.
- Hemben, A.; Ashley, J.; Tothill, I.; Development of an Immunosensor for PfHRP 2 as a Biomarker for Malaria Detection.. Biosensors 2017, , 7, , 28..
- Hemben, A.; Ashley, J.; Tothill, I.E.; An immunosensor for parasite lactate dehydrogenase detection as a malaria biomarker – Comparison with commercial test kit. Talanta 2018, , 187, , 321–329..
- Babina Chakma; Priyamvada Jain; Naveen K. Singh; Pranab Goswami; Development of an Indicator Displacement Based Detection of Malaria Targeting HRP-II as Biomarker for Application in Point-of-Care Settings. Analytical Chemistry 2016, 88, 10316-10321, 10.1021/acs.analchem.6b03315.
- Roderick M. Dirkzwager; Shaolin Liang; Julian Alexander Tanner; Development of Aptamer-Based Point-of-Care Diagnostic Devices for Malaria Using Three-Dimensional Printing Rapid Prototyping. ACS Sensors 2016, 1, 420-426, 10.1021/acssensors.5b00175.
- Lewis Fraser; Andrew B. Kinghorn; Roderick M. Dirkzwager; Shaolin Liang; Yee-Wai Cheung; Bryce Lim; Simon Chi-Chin Shiu; Sze-Lok Marco Tang; Dean Andrew; Joseph Manitta; et al.Jack RichardsJulian Alexander Tanner A portable microfluidic Aptamer-Tethered Enzyme Capture (APTEC) biosensor for malaria diagnosis.. Biosensors and Bioelectronics 2017, 100, 591-596, 10.1016/j.bios.2017.10.001.
- Mukesh K. Sharma; V.K. Rao; Gauri S. Agarwal; Ganga P. Rai; N. Gopalan; Shri Prakash; S. K. Sharma; R Vijayaraghavan; Highly Sensitive Amperometric Immunosensor for Detection of Plasmodium falciparum Histidine-Rich Protein 2 in Serum of Humans with Malaria: Comparison with a Commercial Kit▿. Journal of Clinical Microbiology 2008, 46, 3759-3765, 10.1128/JCM.01022-08.
- Kenry Kenry; Alisha Geldert; Xiao Zhang; Hua Zhang; Chwee Teck Lim; Highly Sensitive and Selective Aptamer-Based Fluorescence Detection of a Malarial Biomarker Using Single-Layer MoS2Nanosheets. ACS Sensors 2016, 1, 1315-1321, 10.1021/acssensors.6b00449.
- M. De Souza Castilho; T. Laube; H. Yamanaka; S. Alegret; M. I. Pividori; Magneto Immunoassays for Plasmodium falciparum Histidine-Rich Protein 2 Related to Malaria based on Magnetic Nanoparticles. Analytical Chemistry 2011, 83, 5570-5577, 10.1021/ac200573s.
- Bhavna Sikarwar; Pushpendra K. Sharma; Anchal Srivastava; Gauri S. Agarwal; Mannan Boopathi; Beer Singh; Yogesh K. Jaiswal; Surface plasmon resonance characterization of monoclonal and polyclonal antibodies of malaria for biosensor applications. Biosensors and Bioelectronics 2014, 60, 201-209, 10.1016/j.bios.2014.04.025.
- Tiparat Potipitak; Warunee Ngrenngarmlert; Chamras Promptmas; Sirinart Chomean; Wanida Ittarat; Diagnosis and genotyping of Plasmodium falciparum by a DNA biosensor based on quartz crystal microbalance (QCM). Clinical Chemistry and Laboratory Medicine (CCLM) 2011, 49, 1367-1373, 10.1515/cclm.2011.178.
- Marianne Smedegaard Hede; Patricia N. Okorie; Signe Kirk Fruekilde; Søren Fjelstrup; Jonas Thomsen; Oskar Franch; Cinzia Tesauro; Magnus Tobias Bugge; Mette Christiansen; Stephane Picot; et al.Felix LötschGhyslain Mombo-NgomaJohannes MischlingerAyôla A. AdegnikaFinn Skou PedersenMegan Yi-Ping HoEskild PetersenMagnus StougaardMichael RamharterBirgitta R. Knudsen Refined Method for Droplet Microfluidics-Enabled Detection of Plasmodium falciparum Encoded Topoisomerase I in Blood from Malaria Patients. Micromachines 2015, 6, 1505-1513, 10.3390/mi6101432.
- Hoan T. Ngo; Naveen Gandra; Andrew M. Fales; Steve M. Taylor; Tuan Vo-Dinh; Sensitive DNA detection and SNP discrimination using ultrabright SERS nanorattles and magnetic beads for malaria diagnostics. Biosensors and Bioelectronics 2016, 81, 8-14, 10.1016/j.bios.2016.01.073.
- Nantawan Wangmaung; Sirinart Chomean; Chamras Promptmas; Sumana Mas-Oodi; Dalina Tanyong; Wanida Ittarat; Silver quartz crystal microbalance for differential diagnosis of Plasmodium falciparum and Plasmodium vivax in single and mixed infection. Biosensors and Bioelectronics 2014, 62, 295-301, 10.1016/j.bios.2014.06.052.
- Binod Kumar; Vijayender Bhalla; Ravi Pratap Singh Bhadoriya; C Raman Suri; Grish C. Varshney; Label-free electrochemical detection of malaria-infected red blood cells. RSC Advances 2016, 6, 75862-75869, 10.1039/C6RA07665C.
- Tian Fook Kong; Weijian Ye; Weng Kung Peng; Han Wei Hou; Marcos Marcos; Peter Rainer Preiser; Anh V. Nguyen; Jongyoon Han; Enhancing malaria diagnosis through microfluidic cell enrichment and magnetic resonance relaxometry detection. Scientific Reports 2015, 5, 1-2, 10.1038/srep11425.
- M Rodriguez-Del Valle; I A Quakyi; J Amuesi; J T Quaye; F K Nkrumah; D W Taylor; Detection of antigens and antibodies in the urine of humans with Plasmodium falciparum malaria.. Journal of Clinical Microbiology 1991, 29, 1236-1242, 10.1128/jcm.29.6.1236-1242.1991.
- M E Parra; C B Evans; D W Taylor; Identification of Plasmodium falciparum histidine-rich protein 2 in the plasma of humans with malaria.. Journal of Clinical Microbiology 1991, 29, 1629-1634, null.
- Mandy L Y Sin; Kathleen E Mach; Pak Kin Wong; Joseph C. Liao; Advances and challenges in biosensor-based diagnosis of infectious diseases.. Expert Review of Molecular Diagnostics 2014, 14, 225-244, 10.1586/14737159.2014.888313.
- Yagahira E. Castro-Sesquen; Chloe Kim; Robert H. Gilman; David J. Sullivan; Peter C. Searson; Nanoparticle-Based Histidine-Rich Protein-2 Assay for the Detection of the Malaria Parasite Plasmodium falciparum. The American Journal of Tropical Medicine and Hygiene 2016, 95, 354-357, 10.4269/ajtmh.15-0772.
- Xiaodong Cao; Yongkang Ye; Songqin Liu; Gold nanoparticle-based signal amplification for biosensing. Analytical Biochemistry 2011, 417, 1-16, 10.1016/j.ab.2011.05.027.
- Jianping Lei; Huangxian Ju; Signal Amplification Using Nanomaterials for Biosensing. Optical Nano- and Microsystems for Bioanalytics 2013, 14, 17-41, 10.1007/5346_2012_46.
- Huangxian Ju; Xueji Zhang; Joseph Wang; Signal Amplification for Nanobiosensing. Computational Methods for Protein Structure Prediction and Modeling 2011, null, 39-84, 10.1007/978-1-4419-9622-0_2.
- Mukesh K. Sharma; Gauri S. Agarwal; Vepa K. Rao; Sanjay Upadhyay; S. Merwyn; Natarajan Gopalan; Ganga P. Rai; R Vijayaraghavan; Shri Prakash; Amperometric immunosensor based on gold nanoparticles/alumina sol–gel modified screen-printed electrodes for antibodies to Plasmodium falciparum histidine rich protein-2. The Analyst 2010, 135, 608, 10.1039/b918880k.
- M.K. Sharma; V.K. Rao; S. Merwyn; G.S. Agarwal; Sanjay Upadhyay; R Vijayaraghavan; A novel piezoelectric immunosensor for the detection of malarial Plasmodium falciparum histidine rich protein-2 antigen. Talanta 2011, 85, 1812-1817, 10.1016/j.talanta.2011.07.008.
- Mamantos Prodromidis; Impedimetric immunosensors—A review. Electrochimica Acta 2010, 55, 4227-4233, 10.1016/j.electacta.2009.01.081.
- Piper, R.; LeBras, J.; Wentworth, L.; Hunt-Cooke, A.; Houzé, S.; Chiodini, P.; Makler, M. Immunocapture diagnostic assays for malaria using Plasmodium lactate dehydrogenase (pLDH). Am. J. Trop. Med. Hyg. 1999, 60, 109–118.
- Palmer, C.J.; Lindo, J.F.; Klaskala, W.I.; Quesada, J.A.; Kaminsky, R.; Baum, M.K.; Ager, A.L. Evaluation of the optimal test for rapid diagnosis of Plasmodium vivax and Plasmodium falciparurn malaria. J. Clin. Microbiol. 1998, 36, 203–206.
- M. T. Makler C. J. Palmer A. L. Ager; A review of practical techniques for the diagnosis of malaria. Annals of Tropical Medicine & Parasitology 1998, 92, 419-433, 10.1080/00034989859401.
- Figueroa-Miranda, G.; Feng, L.; Shiu, S.C.C.; Dirkzwager, R.M.; Cheung, Y.W.; Tanner, J.A.; Schöning, M.J.; Offenhäusser, A.; Mayer, D.; Aptamer-based electrochemical biosensor for highly sensitive and selective malaria detection with adjustable dynamic response range and reusability. Sensors Actuators, B Chem. 2018, , 255, , 235–243..
- Jessica Cohen; Pascaline Dupas; Simone Schaner; Price Subsidies, Diagnostic Tests, and Targeting of Malaria Treatment: Evidence from a Randomized Controlled Trial. American Economic Review 2015, 105, 609-645, 10.1257/aer.20130267.
- Priyamvada Jain; Babina Chakma; Sanjukta Patra; Pranab Goswami; Potential Biomarkers and Their Applications for Rapid and Reliable Detection of Malaria. BioMed Research International 2014, 2014, 1-20, 10.1155/2014/852645.
- K. Linda Britton; Patrick Baker; David W. Rice; Timothy J. Stillman; Structural relationship between the hexameric and tetrameric family of glutamate dehydrogenases. JBIC Journal of Biological Inorganic Chemistry 1992, 209, 851-859, 10.1111/j.1432-1033.1992.tb17357.x.
- David L. Vander Jagt; Clare Intress; John E. Heidrich; John E. K. Mrema; Karl H. Rieckmann; Hans-G. Heidrich; Marker Enzymes of Plasmodium falciparum and Human Erythrocytes as Indicators of Parasite Purity. Journal of Parasitology 1982, 68, 1068, 10.2307/3281093.
- Kathleen Zocher; Karin Fritz-Wolf; Sebastian Kehr; Marina Fischer; Stefan Rahlfs; Katja Becker; Biochemical and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2. Molecular and Biochemical Parasitology 2012, 183, 52-62, 10.1016/j.molbiopara.2012.01.007.
- Yan Li; Yun-Shan Ning; Li Li; Dan-Dan Peng; Wen-Qi Dong; Ming Li; [Preparation of a monoclonal antibodies against Plasmodium falciparum glutamate dehydrogenase and establishment of colloidal gold-immunochromatographic assay].. Di 1 jun yi da xue xue bao = Academic journal of the first medical college of PLA 2005, 25, 435-438..
- Jan T. Wagner; Heike Lüdemann; Petra M. Färber; Friedrich Lottspeich; R. Luise Krauth-Siegel; Glutamate dehydrogenase, the marker protein of Plasmodium falciparum . Cloning, expression and characterization of the malarial enzyme. JBIC Journal of Biological Inorganic Chemistry 1998, 258, 813-819, 10.1046/j.1432-1327.1998.2580813.x.
- Park, S.J.; Kwon, O.S.; Lee, S.H.; Song, H.S.; Park, T.H.; Jang, J.; Ultrasensitive Flexible Graphene Based Field-Effect Transistor (FET)-Type Bioelectronic Nose. Nano Lett. 2012, , 12, , 5082–5090..
- I K Srivastava; M Schmidt; U Certa; H Döbeli; L H Perrin; Specificity and inhibitory activity of antibodies to Plasmodium falciparum aldolase.. The Journal of Immunology 1990, 144, 1497-1503.
- Dzakah, E.E.; Kang, K.; Ni, C.; Tang, S.; Wang, J.; Wang, J.; Comparative performance of aldolase and lactate dehydrogenase rapid diagnostic tests in Plasmodium vivax detection. Malar. J. 2014, , 13, , 272..
- Nelson Lee; Joanne Baker; David Bell; James McCarthy; Qin Cheng; Assessing the Genetic Diversity of the Aldolase Genes of Plasmodium falciparum and Plasmodium vivax and Its Potential Effect on Performance of Aldolase-Detecting Rapid Diagnostic Tests▿. Journal of Clinical Microbiology 2006, 44, 4547-4549, 10.1128/JCM.01611-06.
- Monika Chugh; Vidhya Sundararaman; Saravanan Kumar; V. S. Reddy; W. A. Siddiqui; K. D. Stuart; Pawan Malhotra; Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proceedings of the National Academy of Sciences 2013, 110, 5392-5397, 10.1073/pnas.1218412110.
- Silvina Pagola; Peter W. Stephens; D. Scott Bohle; Andrew D. Kosar; Sara K. Madsen; The structure of malaria pigment β-haematin. Nature 2000, 404, 307-310, 10.1038/35005132.
- Chen, K.; Yuen, C.; Aniweh, Y.; Preiser, P.; Liu, Q.; Towards ultrasensitive malaria diagnosis using surface enhanced Raman spectroscopy. Sci. Rep. 2016, , 6, , 20177..
- Bassem S. S. Guirgis; Claudia Sá E Cunha; Inês Gomes; Miguel Cavadas; Isabel Bernardo Goncalves Oliveira Silva; Gonçalo Doria; Gregory Blatch; Pedro Viana Baptista; Eulália Pereira; Hassan M. E. Azzazy; Maria M. Mota; Miguel Prudêncio; Ricardo Franco; Gold nanoparticle-based fluorescence immunoassay for malaria antigen detection. Analytical and Bioanalytical Chemistry 2011, 402, 1019-1027, 10.1007/s00216-011-5489-y.
- Eline Kattenberg; Inge Versteeg; Stephanie J. Migchelsen; Iveth J. González; Mark D. Perkins; Pètra F. Mens; Henk D.F.H. Schallig; New developments in malaria diagnostics. mAbs 2012, 4, 120-126, 10.4161/mabs.4.1.18529.
- J.P. Dean Goldring; The Roles of Acetic Acid and Methanol During Fixing and Staining Proteins in an SDS–Polyacrylamide Electrophoresis Gel. Advanced Structural Safety Studies 2018, null, 15-18, 10.1007/978-1-4939-8745-0_2.
- Garcia, L.S. Diagnostic Medical Parasitology, 5th ed.; ASM Press: Washington, DC, USA, 2007; pp. 130–140.
- Birch, C.M.; Hou, H.W.; Han, J.; Niles, J.C.; Identification of malaria parasite-infected red blood cell surface aptamers by inertial microfluidic SELEX (I-SELEX). Sci. Rep. 2015, , 5, , 11347. .
- Cardoso, A.R.; Cabral-Miranda, G.; Reyes-Sandoval, A.; Bachmann, M.F.; Sales, M.G.F.; Detecting circulating antibodies by controlled surface modification with specific target proteins: Application to malaria. Biosens. Bioelectron. 2017, , 91, , 833–841..
- Cordray, M.S.; Richards-Kortum, R.R.; Review: Emerging nucleic acid-based tests for point-of-care detection of malaria. Am. J. Trop. Med. Hyg. 2012, , 87, , 223–230..
- Wanida Ittarat; Sirinart Chomean; Chularat Sanchomphu; Nantawan Wangmaung; Chamras Promptmas; Warunee Ngrenngarmlert; Biosensor as a molecular malaria differential diagnosis. Clinica Chimica Acta 2013, 419, 47-51, 10.1016/j.cca.2013.01.010.
- Michael A. Nash; John N. Waitumbi; Allan S. Hoffman; Paul Yager; Patrick S. Stayton; Multiplexed Enrichment and Detection of Malarial Biomarkers Using a Stimuli-Responsive Iron Oxide and Gold Nanoparticle Reagent System. ACS Nano 2012, 6, 6776-6785, 10.1021/nn3015008.
- Christine Markwalter; Keersten M. Ricks; Anna L. Bitting; Lwiindi Mudenda; David W. Wright; Simultaneous capture and sequential detection of two malarial biomarkers on magnetic microparticles. Talanta 2016, 161, 443-449, 10.1016/j.talanta.2016.08.078.
- Micha Phill Grønholm Jepsen; Dennis Röser; M Christiansen; Severin Olesen Larsen; David Cavanagh; Kelwalin Dhanasarnsombut; Ib Bygbjerg; Daniel Dodoo; Edmond J. Remarque; Morten Hanefeld Dziegiel; Søren Jepsen; Benjamin Mordmüller; Michael Theisen; Development and evaluation of a multiplex screening assay for Plasmodium falciparum exposure. Journal of Immunological Methods 2012, 384, 62-70, 10.1016/j.jim.2012.07.009.
- Jamshaid Iqbal; Ahmed Siddique; Mohammad Jameel; Persotum R. Hira; Persistent Histidine-Rich Protein 2, Parasite Lactate Dehydrogenase, and Panmalarial Antigen Reactivity after Clearance of Plasmodium falciparum Monoinfection. Journal of Clinical Microbiology 2004, 42, 4237-4241, 10.1128/JCM.42.9.4237-4241.2004.
- Lisa LaFleur; Dean Stevens; Katherine McKenzie; Sujatha Ramachandran; Paolo Spicar-Mihalic; Mitra Singhal; Amit Arjyal; Jennifer Osborn; Peter Kauffman; Paul Yager; et al.Barry Lutz Progress toward multiplexed sample-to-result detection in low resource settings using microfluidic immunoassay cards. Lab on a Chip 2012, 12, 1119, 10.1039/c2lc20751f.
- Rachel N. Deraney; Charles Mace; Jason P. Rolland; Jeremy E. Schonhorn; Multiplexed, Patterned-Paper Immunoassay for Detection of Malaria and Dengue Fever. Analytical Chemistry 2016, 88, 6161-6165, 10.1021/acs.analchem.6b00854.
