Carbon fixation or сarbon assimilation is the process by which inorganic carbon (particularly in the form of carbon dioxide) is converted to organic compounds by living organisms. The compounds are then used to store energy and as structure for other biomolecules. Carbon is primarily fixed through photosynthesis, but some organisms use a process called chemosynthesis in the absence of sunlight. Organisms that grow by fixing carbon are called autotrophs, which include photoautotrophs (which use sunlight), and lithoautotrophs (which use inorganic oxidation). Heterotrophs are not themselves capable of carbon fixation but are able to grow by consuming the carbon fixed by autotrophs or other heterotrophs. "Fixed carbon", "reduced carbon", and "organic carbon" may all be used interchangeably to refer to various organic compounds.
- biomolecules
- carbon fixation
- inorganic carbon
1. Net Vs. Gross CO2 Fixation
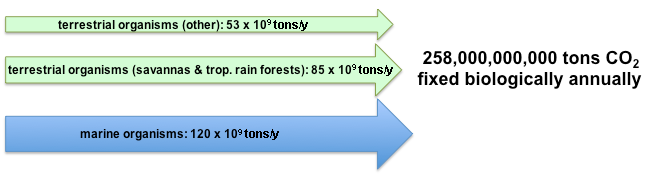
The primary form of inorganic carbon that is fixed is carbon dioxide (CO2). It is estimated that approximately 258 billion tons of carbon dioxide are converted by photosynthesis annually. The majority of the fixation occurs in terrestrial environments, especially the tropics. The gross amount of carbon dioxide fixed is much larger since approximately 40% is consumed by respiration following photosynthesis.[1]
2. Overview of Pathways
While carbon is primarily fixed through the 6 autotrophic pathways, there are non-autotrophic pathways as well.
Six autotrophic carbon fixation pathways are known as of 2011. The Calvin cycle fixes carbon in the chloroplasts of plants and algae, and in the cyanobacteria. It also fixes carbon in the anoxygenic photosynthesis in one type of proteobacteria called purple bacteria, and in some non-phototrophic proteobacteria.[2]
Of the five other autotrophic pathways, two are known only in bacteria (the reductive citric acid cycle and the 3-hydroxypropionate cycle), two only in archaea (two variants of the 3-hydroxypropionate cycle), and one in both bacteria and archaea (the reductive acetyl CoA pathway).
3. Oxygenic Photosynthesis
In photosynthesis, energy from sunlight drives the carbon fixation pathway. Oxygenic photosynthesis is used by the primary producers—plants, algae, and cyanobacteria. They contain the pigment chlorophyll, and use the Calvin cycle to fix carbon autotrophically. The process works like this:
- 2H2O → 4e− + 4H+ + O2
- CO2 + 4e− + 4H+ → CH2O + H2O
In the first step, water is dissociated into electrons, protons, and free oxygen. This allows the use of water, one of the most abundant substances on Earth, as an electron donor—as a source of reducing power. The release of free oxygen is a side-effect of enormous consequence. The first step uses the energy of sunlight to oxidize water to O2, and, ultimately, to produce ATP
- ADP + Pi ⇌ ATP + H2O
and the reductant, NADPH
- NADP+ + 2e− + 2H+ ⇌ NADPH + H+
In the second step, called the Calvin cycle, the actual fixation of carbon dioxide is carried out. This process consumes ATP and NADPH. The Calvin cycle in plants accounts for the preponderance of carbon fixation on land. In algae and cyanobacteria, it accounts for the preponderance of carbon fixation in the oceans. The Calvin cycle converts carbon dioxide into sugar, as triose phosphate (TP), which is glyceraldehyde 3-phosphate (GAP) together with dihydroxyacetone phosphate (DHAP):
- 3 CO2 + 12 e− + 12 H+ + Pi → TP + 4 H2O
An alternative perspective accounts for NADPH (source of e−) and ATP:
- 3 CO2 + 6 NADPH + 6 H+ + 9 ATP + 5 H2O → TP + 6 NADP+ + 9 ADP + 8 Pi
The formula for inorganic phosphate (Pi) is HOPO32− + 2H+. Formulas for triose and TP are C2H3O2-CH2OH and C2H3O2-CH2OPO32− + 2H+
3.1. Evolutionary Considerations
Somewhere between 3.8 and 2.3 billion years ago, the ancestors of cyanobacteria evolved oxygenic photosynthesis,[3][4] enabling the use of the abundant yet relatively oxidized molecule H2O as an electron donor to the electron transport chain of light-catalyzed proton-pumping responsible for efficient ATP synthesis.[5][6] When this evolutionary breakthrough occurred, autotrophy (growth using inorganic carbon as the sole carbon source) is believed to have already been developed. However, the proliferation of cyanobacteria, due to their novel ability to exploit water as a source of electrons, radically altered the global environment by oxygenating the atmosphere and by achieving large fluxes of CO2 consumption.[7]
3.2. CO2 Concentrating Mechanisms
Many photosynthetic organisms have not acquired CO2 concentrating mechanisms (CCMs), which increase the concentration of CO2 available to the initial carboxylase of the Calvin cycle, the enzyme RuBisCO. The benefits of a CCM include increased tolerance to low external concentrations of inorganic carbon, and reduced losses to photorespiration. CCMs can make plants more tolerant of heat and water stress.
CO2 concentrating mechanisms use the enzyme carbonic anhydrase (CA), which catalyze both the dehydration of bicarbonate to CO2 and the hydration of CO2 to bicarbonate
- HCO3− + H+ ⇌ CO2 + H2O
Lipid membranes are much less permeable to bicarbonate than to CO2. To capture inorganic carbon more effectively, some plants have adapted the anaplerotic reactions
- HCO3− + H+ + PEP → OAA + Pi
catalyzed by PEP carboxylase (PEPC), to carboxylate phosphoenolpyruvate (PEP) to oxaloacetate (OAA) which is a C4 dicarboxylic acid.
CAM plants
CAM plants that use Crassulacean acid metabolism as an adaptation for arid conditions. CO2 enters through the stomata during the night and is converted into the 4-carbon compound, malic acid, which releases CO2 for use in the Calvin cycle during the day, when the stomata are closed. The dung jade plant (Crassula ovata) and cacti are typical of CAM plants. Sixteen thousand species of plants use CAM.[8] These plants have a carbon isotope signature of −20 to −10 ‰.[9]
C4 plants
C4 plants preface the Calvin cycle with reactions that incorporate CO2 into one of the 4-carbon compounds, malic acid or aspartic acid. C4 plants have a distinctive internal leaf anatomy. Tropical grasses, such as sugar cane and maize are C4 plants, but there are many broadleaf plants that are C4. Overall, 7600 species of terrestrial plants use C4 carbon fixation, representing around 3% of all species.[10] These plants have a carbon isotope signature of −16 to −10 ‰.[9]
C3 plants
The large majority of plants are C3 plants. They are so-called to distinguish them from the CAM and C4 plants, and because the carboxylation products of the Calvin cycle are 3-carbon compounds. They lack C4 dicarboxylic acid cycles, and therefore have higher CO2 compensation points than CAM or C4 plants. C3 plants have a carbon isotope signature of −24 to −33‰.[9]
Bacteria and cyanobacteria
Almost all cyanobacteria and some bacteria utilize carboxysomes to concentrate carbon dioxide.[11] Carboxysomes are protein shells filled with the enzyme RuBisCO and a carbonic anhydrase. The carbonic anhydrase produces CO2 from the bicarbonate that diffuses into the carboxysome. The surrounding shell provides a barrier to carbon dioxide loss, helping to increase its concentration around RuBisCO.
Eukaryotic algae
In eukaryotic algae, various bicarbonate transporters and carbonic anhydrases [12] serve to increase the CO2 flux balance toward the pyrenoid, a low CO2-permeable subcellular compartment in the chloroplast containing most of the RuBisCO.[13][14]
4. Other Autotrophic Pathways
4.1. Reverse Krebs Cycle
The reverse Krebs cycle, also known as reverse TCA cycle (rTCA) or reductive citric acid cycle, is an alternative to the standard Calvin-Benson cycle for carbon fixation. It has been found in strict anaerobic or microaerobic bacteria (as Aquificales)[15] and anaerobic archea. It was discovered by Evans, Buchanan and Arnon in 1966 working with the photosynthetic green sulfur bacterium Chlorobium limicola.[16] The cycle involves the biosynthesis of acetyl-CoA from two molecules of CO2.[17] The key steps of the reverse Krebs cycle are:
- Oxaloacetate to malate, using NADH + H+
[math]\ce{ Oxaloacetate + NADH/H+ -> Malate + NAD+ }[/math]
- Fumarate to succinate, catalyzed by an oxidoreductase, Fumarate reductase
[math]\ce{ Fumarate + FADH2 <=> Succinate + FAD }[/math]
- Succinate to succinyl-CoA, an ATP dependent step
[math]\ce{ Succinate + ATP + CoA -> Succinyl-CoA + ADP + Pi }[/math]
- Succinyl-CoA to alpha-ketoglutarate, using one molecule of CO2
[math]\ce{ Succinyl-CoA + CO2 + Fd{(red)} -> alpha-ketoglutarate + Fd{(ox)} }[/math]
- Alpha-ketoglutarate to isocitrate, using NADPH + H+ and another molecule of CO2
[math]\ce{ Alpha-ketoglutarate + CO2 + NAD(P)H/H+ -> Isocitrate + NAD(P)+ }[/math]
- Citrate converted into oxaloacetate and acetyl-CoA, this is an ATP dependent step and the key enzyme is the ATP citrate lyase
[math]\ce{ Citrate + ATP + CoA -> Oxaloacetate + Acetyl-CoA + ADP + Pi }[/math]
This pathway is cyclic due to the regeneration of the oxaloacetate.[18]
The reverse Krebs cycle is used by microorganisms in anaerobic environments. In particular, it is one of the most used pathways in hydrothermal vents by the Epsilonproteobacteria.[19] This feature is very important in oceans. Without it, there would be no primary production in aphotic environments, which would lead to habitats without life. So this kind of primary production is called "dark primary production".[20]
One other important aspect is the symbiosis between Gammaproteobacteria and Riftia pachyptila. These bacteria can switch from the Calvin-Benson cycle to the rTCA cycle and vice versa in response to different concentrations of H2S in the environment.[21]
4.2. Reductive Acetyl CoA pathway
The reductive acetyl CoA pathway (CoA) pathway, also known as the Wood-Ljungdahl pathway, was discovered by Harland G. Wood and Lars G. Ljungdahl in 1965, thanks to their studies on Clostridium thermoaceticum, a Gram positive bacterium now named Moorella thermoacetica.[22] It is an acetogen, an anaerobic bacteria that uses CO2 as electron acceptor and carbon source, and H2 as an electron donor to form acetic acid.[23][24][25][26] This metabolism is wide spread within the phylum Firmicutes, especially in the Clostridia.[23]
The pathway is also used by methanogens, which are mainly Euryarchaeota, and several anaerobic chemolithoautotrophs, such as sulfate-reducing bacteria and archaea. It is probably performed also by the Brocadiales, an order of Planctomycetes that oxidize ammonia in anaerobic condition.[17][27][28][29][30][31][32] Hydrogenotrophic methanogenesis, which is only found in certain archaea and accounts for 80% of global methanogenesis, is also based on the reductive acetyl CoA pathway.
The Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase is the oxygen-sensitive enzyme that permits the reduction of CO2 to CO and the synthesis of acetyl-CoA in several reactions.[33]
One branch of this pathway, the methyl branch, is similar but non-homologous between bacteria and archaea. In this branch happens the reduction of CO2 to a methyl residue bound to a cofactor. The intermediates are formate for bacteria and formyl-methanofuran for archaea, and also the carriers, tetrahydrofolate and tetrahydropterins respectively in bacteria and archaea, are different, such as the enzymes forming the cofactor-bound methyl group.[17]
Otherwise, the carbonyl branch is homologous between the two domains and consists of the reduction of another molecule of CO2 to a carbonyl residue bound to an enzyme, catalyzed by the CO dehydrogenase/acetyl-CoA synthase. This key enzyme is also the catalyst for the formation of acetyl-CoA starting from the products of the previous reactions, the methyl and the carbonyl residues.[33][34]
This carbon fixation pathway requires only one molecule of ATP for the production of one molecule of pyruvate, which makes this process one of the main choice for chemolithoautotrophs limited in energy and living in anaerobic conditions.[17]
4.3. 3-Hydroxypropionate Bicycle
The 3-Hydroxypropionate bicycle, also known as 3-HP/malyl-CoA cycle, was discovered by Helge Holo in 1989. It's a pathway of carbon fixation and is utilized by green non-sulfur phototrophs of Chloroflexaceae family, including the maximum exponent of this family Chloroflexus auranticus by which this way was discovered and demonstrated.[35]
The 3-Hydroxipropionate bicycle is composed of two cycles and the name of this way comes from the 3-Hydroxyporopionate which corresponds to an intermediate characteristic of it.
The first cycle is a way of synthesis of glycoxilate. During this cycle two bicarbonate molecules are fixed thanks to the action of two enzymes: the Acetyl-CoA carboxylase catalyzes the carboxylation of the Acetyl-CoA to Malonyl-CoA and Propionyl-CoA carboxylase catalyses the carboxylation of Propionyl-CoA to Methylamalonyl-CoA. From this point a series of reactions lead to the formation of glycoxylate which will thus become part of the second cycle.[36][37]
In the second cycle, glycoxilate is approximately one molecule of Propionyl-CoA forming Methylamalonyl-CoA. This, in turn, is then converted through a series of reactions into Citramalyl-CoA. The Citramalyl-CoA is split into pyruvate and Acetyl-CoA thanks to the enzyme MMC lyase. At this point the pyruvate is released, while the Acetyl-CoA is reused and carboxylated again at Malonyl-coa thus reconstituting the cycle.[38]
19 are the total reactions involved in 3-Hydroxypropionate bicycle and 13 are the multifunctional enzymes used. The multifunctionality of these enzymes is an important feature of this pathway which thus allows the fixation of 3 bicarbonate molecules.[38]
It is a very expensive way: 7 ATP molecules are used for the synthesis of the new pyruvate and 3 ATP for the phosphate triose.[37]
An important characteristic of this cycle is that it allows the co-assimilation of numerous compounds making it suitable for the mixotrophic organisms.[37]
A variant of the 3-hydroxypropionate cycle was found to operate in the aerobic extreme thermoacidophile archaeon Metallosphaera sedula. This pathway is called the 3-hydroxypropionate/4-hydroxybutyrate cycle.[39]
Yet another variant of the 3-hydroxypropionate cycle is the dicarboxylate/4-hydroxybutyrate cycle. It was discovered in anaerobic archaea. It was proposed in 2008 for the hyperthermophile archeon Ignicoccus hospitalis.[40]
5. Chemosynthesis
Chemosynthesis is carbon fixation driven by energy obtained by oxidating inorganic substances (e.g., hydrogen gas or hydrogen sulfide), rather than from sunlight. Sulfur- and hydrogen-oxidizing bacteria often use the Calvin cycle or the reductive citric acid cycle.[41]
6. Non-Autotrophic Pathways
Although almost all heterotrophs cannot synthesize complete organic molecules from carbon dioxide, some carbon dioxide is incorporated in their metabolism.[42] Notably pyruvate carboxylase consumes carbon dioxide (as bicarbonate ions) as part of gluconeogenesis, and carbon dioxide is consumed in various anaplerotic reactions. Recently, also 6-phosphogluconate dehydrogenase was shown to catalyze the reductive carboxylation of ribulose 5-phosphate to 6-phosphogluconate in E. coli under elevated CO2 concentrations.[43] Considering the CO2 concentration in the habitat of E. coli (e.g. the mammalian gut[44]), this reaction might happen also naturally. In the future, this property could be exploited for the design of synthetic carbon fixation routes.
7. Carbon Isotope Discrimination
Some carboxylases, particularly RuBisCO, preferentially bind the lighter carbon stable isotope carbon-12 over the heavier carbon-13. This is known as carbon isotope discrimination and results in carbon-12 to carbon-13 ratios in the plant that are higher than in the free air. Measurement of this ratio is important in the evaluation of water use efficiency in plants,[45][46][47] and also in assessing the possible or likely sources of carbon in global carbon cycle studies.
References
- "Primary productivity of planet earth: biological determinants and physical constraints in terrestrial and aquatic habitats". Global Change Biology 7 (8): 849–882. 2001. doi:10.1046/j.1365-2486.2001.00448.x. Bibcode: 2001GCBio...7..849G. https://dx.doi.org/10.1046%2Fj.1365-2486.2001.00448.x
- "Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean". Science 333 (6047): 1296–300. September 2011. doi:10.1126/science.1203690. PMID 21885783. Bibcode: 2011Sci...333.1296S. https://dx.doi.org/10.1126%2Fscience.1203690
- "Early Archean origin of Photosystem II". Geobiology 17 (2): 127–150. March 2019. doi:10.1111/gbi.12322. PMID 30411862. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=6492235
- "Origin and Evolution of Water Oxidation before the Last Common Ancestor of the Cyanobacteria". Molecular Biology and Evolution 32 (5): 1310–28. May 2015. doi:10.1093/molbev/msv024. PMID 25657330. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4408414
- "A fresh look at the fossil evidence for early Archaean cellular life". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 361 (1470): 887–902. June 2006. doi:10.1098/rstb.2006.1835. PMID 16754605. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1578727
- "The evolutionary diversification of cyanobacteria: molecular-phylogenetic and paleontological perspectives". Proceedings of the National Academy of Sciences of the United States of America 103 (14): 5442–7. April 2006. doi:10.1073/pnas.0600999103. PMID 16569695. Bibcode: 2006PNAS..103.5442T. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1459374
- "The Paleoproterozoic snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis". Proceedings of the National Academy of Sciences of the United States of America 102 (32): 11131–6. August 2005. doi:10.1073/pnas.0504878102. PMID 16061801. Bibcode: 2005PNAS..10211131K. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1183582
- "Crassulacean acid metabolism: plastic, fantastic". Journal of Experimental Botany 53 (369): 569–80. April 2002. doi:10.1093/jexbot/53.369.569. PMID 11886877. https://dx.doi.org/10.1093%2Fjexbot%2F53.369.569
- O'Leary MH (1988). "Carbon isotopes in photosynthesis". BioScience 38 (5): 328–336. doi:10.2307/1310735.null
- "16. The Taxonomic Distribution of C4 Photosynthesis". C4 Plant Biology. 1999. pp. 551–580. ISBN 0-12-614440-0.
- Badger, M. R. (2003-02-01). "CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution". Journal of Experimental Botany 54 (383): 609–622. doi:10.1093/jxb/erg076. PMID 12554704. https://dx.doi.org/10.1093%2Fjxb%2Ferg076
- Pierella Karlusich, Juan José; Bowler, Chris; Biswas, Haimanti (2021-04-30). "Carbon Dioxide Concentration Mechanisms in Natural Populations of Marine Diatoms: Insights From Tara Oceans". Frontiers in Plant Science 12: 657821. doi:10.3389/fpls.2021.657821. ISSN 1664-462X. PMID 33995455. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=8119650
- Hennacy, Jessica H.; Jonikas, Martin C. (2020-04-29). "Prospects for Engineering Biophysical CO 2 Concentrating Mechanisms into Land Plants to Enhance Yields" (in en). Annual Review of Plant Biology 71 (1): 461–485. doi:10.1146/annurev-arplant-081519-040100. ISSN 1543-5008. PMID 32151155. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=7845915
- Meyer, Moritz T; Whittaker, Charles; Griffiths, Howard (2017-06-22). "The algal pyrenoid: key unanswered questions" (in en). Journal of Experimental Botany 68 (14): 3739–3749. doi:10.1093/jxb/erx178. ISSN 0022-0957. PMID 28911054. http://academic.oup.com/jxb/article/68/14/3739/3858331/The-algal-pyrenoid-key-unanswered-questions.
- "Before enzymes and templates: theory of surface metabolism.". Microbiological Reviews (American Society for Microbiology) 52 (4): 452–484. 1988. doi:10.1128/mr.52.4.452-484.1988. OCLC 680443998. PMID 3070320. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=373159
- "Alternative pathways of carbon dioxide fixation: insights into the early evolution of life?". Annual Review of Microbiology 65 (1): 631–58. 2011-10-13. doi:10.1146/annurev-micro-090110-102801. PMID 21740227. https://dx.doi.org/10.1146%2Fannurev-micro-090110-102801
- "Beyond the Calvin cycle: autotrophic carbon fixation in the ocean". Annual Review of Marine Science 3 (1): 261–89. 2011-01-15. doi:10.1146/annurev-marine-120709-142712. PMID 21329206. Bibcode: 2011ARMS....3..261H. https://dx.doi.org/10.1146%2Fannurev-marine-120709-142712
- "A reverse KREBS cycle in photosynthesis: consensus at last". Photosynthesis Research 24 (1): 47–53. April 1990. doi:10.1007/bf00032643. PMID 24419764. https://dx.doi.org/10.1007%2Fbf00032643
- "Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility". Proceedings of the National Academy of Sciences of the United States of America 105 (45): 17516–21. November 2008. doi:10.1073/pnas.0802782105. PMID 18987310. Bibcode: 2008PNAS..10517516G. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2579889
- Is dark carbon fixation relevant for oceanic primary production estimates?. 2019-06-11. doi:10.5194/bg-2019-223. https://www.biogeosciences-discuss.net/bg-2019-223/bg-2019-223.pdf.
- "Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila". Science 315 (5809): 247–50. January 2007. doi:10.1126/science.1132913. OCLC 655249163. PMID 17218528. Bibcode: 2007Sci...315..247M. https://dx.doi.org/10.1126%2Fscience.1132913
- "Incorporation of C14 from Carbon Dioxide into Sugar Phosphates, Carboxylic Acids, and Amino Acids by Clostridium thermoaceticum". Journal of Bacteriology 89 (4): 1055–64. April 1965. doi:10.1128/jb.89.4.1055-1064.1965. PMID 14276095. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=277595
- "Old acetogens, new light". Annals of the New York Academy of Sciences 1125 (1): 100–28. March 2008. doi:10.1196/annals.1419.016. PMID 18378590. Bibcode: 2008NYASA1125..100D. https://dx.doi.org/10.1196%2Fannals.1419.016
- "Total synthesis of acetate from CO2 by heterotrophic bacteria". Annual Review of Microbiology 23 (1): 515–38. 1969. doi:10.1146/annurev.mi.23.100169.002503. PMID 4899080. https://dx.doi.org/10.1146%2Fannurev.mi.23.100169.002503
- "The autotrophic pathway of acetate synthesis in acetogenic bacteria". Annual Review of Microbiology 40 (1): 415–50. 1986-01-01. doi:10.1146/annurev.micro.40.1.415. PMID 3096193. https://dx.doi.org/10.1146%2Fannurev.micro.40.1.415
- "A life with acetogens, thermophiles, and cellulolytic anaerobes". Annual Review of Microbiology 63 (1): 1–25. 2009. doi:10.1146/annurev.micro.091208.073617. PMID 19575555. https://dx.doi.org/10.1146%2Fannurev.micro.091208.073617
- "Carbon assimilation pathways in sulfate reducing bacteria. Formate, carbon dioxide, carbon monoxide, and acetate assimilation by Desulfovibrio baarsii". Archives of Microbiology 138 (3): 257–262. 1984. doi:10.1007/bf00402132. ISSN 0302-8933. https://dx.doi.org/10.1007%2Fbf00402132
- "Single-carbon chemistry of acetogenic and methanogenic bacteria". Science 227 (4691): 1167–73. March 1985. doi:10.1126/science.3919443. PMID 3919443. Bibcode: 1985Sci...227.1167Z. https://dx.doi.org/10.1126%2Fscience.3919443
- "Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum". Archives of Microbiology 151 (1): 84–89. 1989. doi:10.1007/bf00444674. https://dx.doi.org/10.1007%2Fbf00444674
- "Variations of the Acetyl-CoA Pathway in Diversely Related Microorganisms That Are Not Acetogens". Acetogenesis. Springer US. 1994. pp. 507–520. doi:10.1007/978-1-4615-1777-1_19. ISBN 978-1-4613-5716-2. https://dx.doi.org/10.1007%2F978-1-4615-1777-1_19
- "Enzymes and coenzymes of the carbon monoxide dehydrogenase pathway for autotrophic CO2 fixation in Archaeoglobus lithotrophicus and the lack of carbon monoxide dehydrogenase in the heterotrophic A. profundus". Archives of Microbiology 163 (2): 112–118. 1995-02-01. doi:10.1007/s002030050179. ISSN 0302-8933. https://dx.doi.org/10.1007%2Fs002030050179
- "Deciphering the evolution and metabolism of an anammox bacterium from a community genome". Nature 440 (7085): 790–4. April 2006. doi:10.1038/nature04647. PMID 16598256. Bibcode: 2006Natur.440..790S. https://dx.doi.org/10.1038%2Fnature04647
- "Role of carbon monoxide dehydrogenase in the autotrophic pathway used by acetogenic bacteria". Proceedings of the National Academy of Sciences of the United States of America 81 (20): 6261–5. October 1984. doi:10.1073/pnas.81.20.6261. PMID 6436811. Bibcode: 1984PNAS...81.6261P. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=391903
- "Acetate biosynthesis by acetogenic bacteria. Evidence that carbon monoxide dehydrogenase is the condensing enzyme that catalyzes the final steps of the synthesis". The Journal of Biological Chemistry 260 (7): 3970–7. April 1985. doi:10.1016/S0021-9258(18)89217-1. PMID 2984190. https://dx.doi.org/10.1016%2FS0021-9258%2818%2989217-1
- "Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle". European Journal of Biochemistry 215 (3): 633–43. August 1993. doi:10.1111/j.1432-1033.1993.tb18074.x. PMID 8354269. https://dx.doi.org/10.1111%2Fj.1432-1033.1993.tb18074.x
- "L-Malyl-coenzyme A lyase/beta-methylmalyl-coenzyme A lyase from Chloroflexus aurantiacus, a bifunctional enzyme involved in autotrophic CO(2) fixation". Journal of Bacteriology 184 (21): 5999–6006. November 2002. doi:10.1128/jb.184.21.5999-6006.2002. PMID 12374834. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=135395
- "Ecological aspects of the distribution of different autotrophic CO2 fixation pathways". Applied and Environmental Microbiology 77 (6): 1925–36. March 2011. doi:10.1128/aem.02473-10. PMID 21216907. Bibcode: 2011ApEnM..77.1925B. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3067309
- "Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus". Proceedings of the National Academy of Sciences of the United States of America 106 (50): 21317–22. December 2009. doi:10.1073/pnas.0908356106. PMID 19955419. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2795484
- "A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea". Science 318 (5857): 1782–6. December 2007. doi:10.1126/science.1149976. PMID 18079405. Bibcode: 2007Sci...318.1782B. https://dx.doi.org/10.1126%2Fscience.1149976
- "A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis". Proceedings of the National Academy of Sciences of the United States of America 105 (22): 7851–6. June 2008. doi:10.1073/pnas.0801043105. PMID 18511565. Bibcode: 2008PNAS..105.7851H. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2409403
- Encyclopedia of Microbiology. Academic Press. 2009. pp. 83–84. ISBN 9780123739445. https://books.google.com/books?id=rLhdW5YzuO4C&q=chemosynthesis+carbon+fixation&pg=RA2-PA83.
- Nicole Kresge; Robert D. Simoni; Robert L. Hill (2005). "The Discovery of Heterotrophic Carbon Dioxide Fixation by Harland G. Wood". The Journal of Biological Chemistry 280 (18): e15. http://www.jbc.org/content/280/18/e15.full.
- "Awakening a latent carbon fixation cycle in Escherichia coli". Nature Communications 11 (1): 5812. November 2020. doi:10.1038/s41467-020-19564-5. PMID 33199707. Bibcode: 2020NatCo..11.5812S. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=7669889
- "Volume and composition of human intestinal gas determined by means of an intestinal washout technic". The New England Journal of Medicine 284 (25): 1394–8. June 1971. doi:10.1056/nejm197106242842502. PMID 5578321. http://dx.doi.org/10.1056/nejm197106242842502.
- "Genetic control of water use efficiency and leaf carbon isotope discrimination in sunflower (Helianthus annuus L.) subjected to two drought scenarios". PLOS ONE 9 (7): e101218. 3 July 2014. doi:10.1371/journal.pone.0101218. PMID 24992022. Bibcode: 2014PLoSO...9j1218A. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4081578
- "Carbon Isotope Discrimination and Photosynthesis". Annual Review of Plant Physiology and Plant Molecular Biology 40 (1): 503–537. June 1989. doi:10.1146/annurev.pp.40.060189.002443. https://dx.doi.org/10.1146%2Fannurev.pp.40.060189.002443
- "Carbon isotopes and water use efficiency: sense and sensitivity". Oecologia 155 (3): 441–54. March 2008. doi:10.1007/s00442-007-0932-7. PMID 18224341. Bibcode: 2008Oecol.155..441S. https://dx.doi.org/10.1007%2Fs00442-007-0932-7
