Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Tong Zhang and Version 4 by Beatrix Zheng.
Protein S-glutathionylation (SSG) is a reversible post-translational modification (PTM) featuring the conjugation of glutathione to a protein cysteine thiol. SSG can alter protein structure, activity, subcellular localization, and interaction with small molecules and other proteins. Thus, it plays a critical role in redox signaling and regulation in various physiological activities and pathological events. Many approaches have been developed for the detection of SSG, including direct detection, selective reduction and tagging approaches, and chemoselective probe-based approaches. Utilization of these methods in profiling the SSG proteome had been reported in various biological systems.
- redox
- redox proteomics
- post-translational modification
1. Direct Detection
Radiolabeled (35S-) GSH was one of the earliest tools used for direct detection of S-glutathionylation (SSG) [1][2][3][20,57,58]. To analyze in vivo SSG, protein synthesis is first blocked by pre-incubating cells with cycloheximide and followed by supplement of 35S-cysteine in cell culture to radioactively label the intracellular GSH pool. Induced by oxidative stress, the synthesized 35S-GSH is employed to form radiolabeled SSG, which can be extracted and visualized on a non-reducing SDS-PAGE gel using autoradiography and phosphor imaging technology (after drying the gel or transferring to nitrocellulose). Using this method, a number of proteins were identified as modified by GSH in vivo, such as GAPDH [4][59], actin [5][60], protein kinase C-α [6][61], and thioredoxin [1][20]. This method was further coupled with peptide mass fingerprinting to identify more proteins in mammalian cells, yeast, and algae [2][3][7][57,58,62]. However, this method has several limitations, including the disturbed cell physiology resulting from the need to block protein synthesis, potential non-specific signals (e.g., radiolabeled cysteinylated proteins), and the inability to distinguish individual SSG sites.
Another type of popular tool for direct detection of SSG is through Western immunoblotting of glutathionylated proteins on one-dimensional (1D) or two-dimensional (2D) non-reducing gels using anti-GSH antibodies [8][9][63,64]. Similar to radiolabeled GSH, the antibody method has limited specificity and sensitivity and is unable to distinguish individual SSG sites within a target protein, which may have different functional consequences. Nevertheless, this method proves to be a useful tool to evaluate global SSG level changes. In conjunction with protein-specific antibodies, it also allows for the quantification of SSG of targeted proteins in complex samples [10][11][12][65,66,67].
Besides the radiolabeled and antibody approaches, mass spectrometry can also directly detect the SSG moiety on proteins or peptides. For example, top-down proteomics has been applied to identify sarcomeric protein PTMs, including SSG [13][14][68,69]. By detecting the glutathione conjugation (+305.03 Da) on proteoforms, SSG on troponin I (TnI) was identified as a potential marker that increases with aging. Alternatively, thwe researchers rececently applied bottom-up MS-based global proteome profiling to directly detect thiol PTMs without enrichment [15][70]. In this approach, proteins were extracted by blocking free thiols and polysulfides (while other thiol PTMs were preserved) and then digested. The resulting peptides were then subjected to fractionation and proteome profiling. In the end, only 81 unique Cys sites with SSG were identified, illustrating the challenge of direct SSG detection by MS proteomics.
2. Selective Reduction and Tagging Approaches
Given the challenges in direct detection of SSG, alternative methods are necessary to enable effective characterization. The selective reduction and tagging strategy has become the most applied approach for enabling enrichment and SSG measurement. This strategy was largely built upon the concept of differential alkylation, which was first introduced in the biotin switch technique (BST) for protein SNO detection [16][71]. BST was later modified to profile other redox PTMs including SSG [17][72]. The reversibility of the target PTMs is the key to this concept. The specifically reduced and tagged proteins or peptides can be subsequently enriched, thus improving the overall coverage of modified proteins significantly. Common concerns of the selective reduction and tagging strategy are potential false identifications caused by potential incomplete alkylation of free thiols and non-specific reduction of targeted PTMs. Evaluation of these critical steps should be considered during method development and optimization.
To selectively target SSG, highly specific glutaredoxin (Grx), such as Grx1C14S and Grx3C14S, C65Y from E. coli, has been used for the deglutathionylation reaction [17][18][19][72,73,74]. Lind et al. first employed the modified BST to enrich and detect SSG specifically reduced by the mutant Grx3 in the presence of GSH, NADPH, and glutathione reductase (GR) [17][72]. Free thiols are first alkylated by NEM, followed by the reduction of SSG by Grx3. The resulting nascent free thiols are then alkylated using NEM-biotin, which can be captured by avidin affinity chromatography (Figure 14A). The captured proteins can be eluted, separated, and visualized on a gel, and further processed for MS-based proteomics analysis. Using this method, 43 novel SSG protein targets were identified including chaperones, cytoskeletal proteins, cell-cycle regulators, and metabolic enzymes in cultured human endothelial-like cells [17][72]. In addition, this method also allowed visualization of SSG in fixed cells by using N-(3-maleimidylpropionyl) biocytin and streptavidin-FITC [20][75]. However, these biotin-based methods were generally ineffective in site-specific SSG identifications.
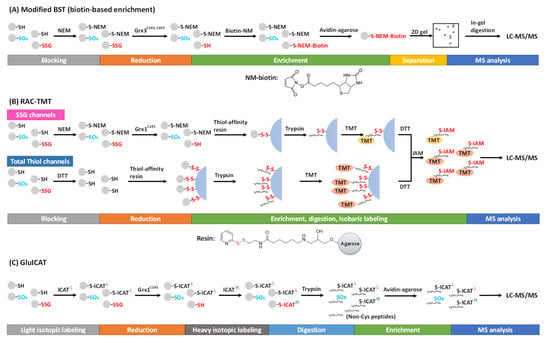
Figure 14. Proteomic approaches for SSG detection and quantification using the selective reduction and tagging strategy. (A) Modified BST, (B) RAC-TMT. and (C) GluICAT approches.
Alternatively, the Qian group introduced a resin-assisted capture (RAC) technique for enriching thiol-containing proteins or peptides [21][22][50,76], which was later adapted for SSG proteome profiling [18][23][24][8,73,77]. In this workflow, following the blocking of free thiols by NEM, protein SSG is selectively reduced by Grx1C14S in the presence of GSSG, NADPH, and GR. The nascent protein thiols are then captured by a thiol-affinity resin, which contains a 2-thiopyridyl disulfide functional group to react with thiol-containing proteins via thiol-disulfide exchange, forming a mixed disulfide bond. The captured proteins can be eluted by DTT for Western blot or proteomics analyses. The stable covalent capture allows for on-resin protein digestion and subsequent multiplexed isobaric labeling (e.g., with tandem mass tags, TMT). Then, the TMT-labeled, Cys-containing peptides can be eluted from the resin by DTT (Figure 14B). This RAC-TMT approach offers several notable advantages. First, the covalent binding of Cys-containing proteins or peptides on the resin permits harsh washing conditions, thus allowing a high enrichment specificity (i.e., typically >95% of final peptides containing cysteines). Second, the multiplex quantification design enables not only comparative quantification of SSG across different biological samples, but also stoichiometric estimation of SSG occupancy at the Cys site level through incorporating both SSG channels and total thiol channels in the same TMT plex experiment. The total thiol samples are prepared with the reduction of all reversibly oxidized thiols by DTT (without initial blocking of the free thiols), thus representing the sum of all thiols on each given Cys site (except irreversible modifications such as SO2H and SO3H) (Figure 14B). Thus, the SSG occupancy at each Cys site can be calculated based on the ratio of TMT reporter ion intensities from the SSG channels to those of the total thiol channels [24][25][77,78]. The quantification of individual post-translational modification (PTM)PTM occupancy at the site-specific level is a critical aspect to understand potential functional consequences of PTMs, which was often overlooked in many current studies. Interested readers are redirected to previous reviews on stoichiometric quantification in redox proteomics [26][27][79,80].
RAC-TMT and similar approaches have been applied to investigate protein SSG changes under physiological and pathological conditions in different cell types and tissues [23][24][25][28][29][8,77,78,81,82]. For example, in a recent study of redox regulation in hyperoxia-induced lung injury, over 7600 SSG sites were quantified by RAC-TMT, providing a landscape view of the SSG proteome in the mouse lung [30][83]. In another study, Duan et al. quantified both SSG and total oxidation occupancies for ~4000 Cys sites under basal conditions in mouse RAW macrophages, revealing a mean occupancy of 4.0% for SSG and 11.9% for total oxidation [25][78]. Interestingly, it was observed that the average occupancies of SSG and total oxidation for individual subcellular compartments were well correlated with their redox potentials.
Recently, an approach called GluICAT, a modified version of the original oxICAT method [31][84], also demonstrated stoichiometric quantification of SSG at the site level [32][33][85,86]. Using isotopically light or heavy thiol-reactive ICAT, free thiols and Grx1C14S-reduced protein SSG are differentially labeled, enriched, and quantified to determine SSG level vs. free thiol level (heavy-to-light ratio) of individual Cys sites (Figure 14C). This method was able to identify 2133 SSG-peptides (~2300 SSG sites) in Leber’s hereditary optic neuropathy (LHON) fibroblasts, where 439 peptides were quantified to show an increased SSG level [32][85]. One notable limitation of this method is that it quantifies the ratio of SSG vs. free thiols without considering other types of thiol PTMs (e.g., disulfides). Thus, the data need to be carefully interpreted with consideration of this important caveat.
Finally, there are several other approaches including the Cys-reactive phosphate tag (CPT)- TMT [34][87] or OxiTMT methods [35][88] that are potentially applicable, but have not yet been demonstrated for SSG analyses.
3. Chemoselective Probes-Based Approaches
The concept of using chemoselective probes to label free thiols either in vivo or in vitro to form protein-SSG analogs is an interesting chemical biology strategy for enabling the characterization of protein-SSG products. In this strategy, glutathione analogs or tagged amino acids (to produce tagged GSH endogenously) were introduced either for in vivo reactions by using membrane permeable probes during cell culture or for in vitro reactions. Glutathione ethyl ester (BioGEE) [36][89] and GSSG-biotin [37][38][90,91] are the first class of glutathione analogs for in vivo and in vitro labeling, and the biotin tag facilitates the enrichment of protein-SSG products. The use of BioGEE or GSSG-biotin is usually accompanied by addition of pro-oxidants to induce oxidative stress, and the resulting protein-SSG products can be identified by Western blot using anti-biotin antibodies or streptavidin-horseradish peroxidase (HRP). The biotin tags also allow capture of glutathionylated proteins by streptavidin agarose beads, which can be released from beads by a reducing reagent (e.g., DTT), thereby cleaving the mixed disulfide bond within the SSG moiety. The eluted proteins can be further analyzed by gel or mass spectrometry [37][39][90,92]. Furthermore, Brennan et al. demonstrated the capacity to image biotinylated protein SSG stained with ExtrAvidin-FITC in cells using confocal fluorescence microscopy [38][91].
Metabolic labeling with clickable GSH probes has been developed as an alternative approach instead of biotin tags. Clickable GSH [40][41][93,94], including azido-GSH, allyl-GSH, and allyl-O-GSH, can be biosynthesized in vivo by expressing an engineered glutathione synthetase (GS M4, F152A/S151G), which catalyzes azido-Ala, allyl-Gly, and allyl-Ser respectively instead of the endogenous substrate Gly (Figure 25). A subsequent click reaction (azide-alkyne or tetrazine-alkene bioorthogonal chemistry) can then tag the glutathionylated proteins with biotin or a fluorophore after protein extraction, thus enabling biotin-streptavidin pull-down or fluorescence labeling for detection. This method has been further extended to LC-MS/MS profiling of glutathionylated proteins by using a cleavable biotin-DADPS-alkyne. Biotinylated proteins are enriched by streptavidin agarose and digested by trypsin, then released by an acidic cleavage of the DADPS linker for detection by LC-MS/MS (Figure 25) [42][95]. By developing light or heavy isotopic labeled clickable GSH, this method was applied to mouse cardiomyocyte cells which quantified 1398 SSG-peptides induced by H2O2 [43][96].
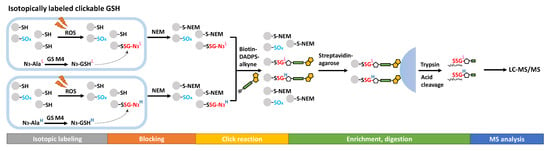
Figure 25.
Proteomics approaches for SSG detection using clickable analogs of GSH.
It should be noted that the addition of GSSG-biotin to cells in culture itself causes oxidative stress due to an increase in cellular GSSG [38][91]. On the other hand, intracellularly synthesized clickable GSH may not disturb the cellular GSH/GSSG ratio. Azido-GSH, azido-GSSG, and an azido-GSH-modified model peptide were reported to be efficient substrates of Grx, GR, and GST omega, which does not interfere with the reversibility of the glutathionylation process and GSH/GSSG cycling [40][93]. However, the application of such probes is limited to living cells that can exogenously express GS M4 and not other types of biological samples, such as tissues. Although the use of chemoselective probes overcomes the potential non-specificity issue of selective reduction and tagging approaches, there are caveats to consider. Introducing these probes themselves is likely to perturb the physiological state and native protein-SSGs are not analyzed.
4. Global Profiling of the SSG Proteome
Enabled by the advances described above, extensive studies for profiling the SSG proteome in various cells or tissues have been reported. Table 1 lists selected examples of recent studies aiming at the global profiling of the SSG proteome, illustrating the different levels of proteome coverage achieved in various samples using different approaches.
Table 1.
Selected MS-based profiling studies of the SSG proteome.
| Biological System | Method | Coverage of the SSG Proteome | Experiment Condition | Significance | Ref. |
|---|---|---|---|---|---|
| Mouse lung | RAC-TMT | ~7600 SSG sites. | Hyperoxia vs. basal; Neonatal wild-type vs. overexpression (β-ENaC) |
Landscape view of SSG-modified proteome in mouse lung and the impact of hypoxia on the SSG proteome | [30][83] |
| RAW 264.7 mouse macrophages | RAC-TMT | Occupancy for 4099 SSG sites | Basal condition | Proteome-wide quantification of both SSG and total oxidation occupancy under physiological conditions, revealing cellular compartmentation of redox homeostasis. | [25][78] |
| Mouse skeletal muscle | RAC-TMT | Occupancy for >2200 SSG sites and changes due to muscle fatiguing | Gastrocnemius muscle with and without fatiguing contractions | Increased muscle protein SSGs identified following a single bout of fatiguing contraction | [24][77] |
| HL-1 cardiomyocyte | Clickable GSH | 1398 SSG-peptides in isotopic duplex experiment | H2O2 treatment (1 mM) | In vivo isotopic tagging of protein SSGs for direct enrichment and quantitative detection; Validation (by Western blot and site mutation) of two structural proteins of interest. | [43][96] |
| Mouse liver | Modified RAC-TMT | 724 SSG-modified proteins | Basal condition, GSTP-nulled mice | The SSG proteome mediated enzymatically by GSTP | [28][81] |
| Synechocystis sp. PCC6803 | GSSG-Biotin | 349 proteins with SSG (protein level enrichment); 145 SSG sites (peptide level enrichment) |
Lysate treated with GSSG-biotin | First SSG proteome profiling in cyanobacteria by GSSG-biotin and LC-MS/MS. | [44][97] |
| Streptococcus mutans UA159 | IodoTMT switch strategy | 357 SSG sites | Wild-type vs. mutants | SSG profiling in in bacteria and mutants; Site mutagenesis validation for SSG function on a thioredoxin-like protein. | [45][98] |
| Human skin fibroblasts | GluICAT | 2307 SSG sites | LHON patients vs. healthy controls | Quantify the ratio of SSG and free thiols with heavy or light ICAT | [32][85] |
