You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 3 by Catherine Yang and Version 2 by Catherine Yang.
miR-21, one of the best-characterized miRNAs to date, has received much attention in renal physiology in particular given its high degree of conservation and expression in kidneys, as well as its potent pathogenic role in various debilitating renal diseases. In contrast with normal kidney function, miR-21 switches to a powerful and overactive mediator under stress conditions. In particular, miR-21 is one of the most highly upregulated miRNAs in a wide panel of tissue injuries, and may act as a cellular sensor of injuries that mediates tissue regeneration.
- microRNA
- kidney
- cancer
- fibrosis
1. miR-21 in Acute and Chronic Kidney Diseases
miR-21 has been associated with the development of a large number of both acute and chronic renal diseases. These studies consistently report a ubiquitous and non-specific increase of miR-21 renal expression in both acute and chronic renal diseases. Numerous experimental models have also explored the role of miR-21 using animal models of acute renal failure or chronic kidney diseases.
1.1. A Protective Role of miR-21 in Acute Kidney Injury?
Acute renal diseases are mainly represented by acute tubular necrosis (ATN), whose causes, although variable, are often related to two main mechanisms: ischemia (induced by hypovolemia, hemorrhage...) and iatrogeny (aminoglycosides, iodine, renin-angiotensin system blockers, cisplatin...) [1]. The evaluation of renal miR-21 expression in clinical samples remains patchy, due to the fact that renal biopsies are rarely performed, except for in the context of renal transplantation [2]. ATN is mainly associated with an increased miR-21 level in renal tissue, serum, or urine of patients with ATN [2][3][4][5]. Furthermore, a large number of ATN-mimicking animal models rely on ischemia-reperfusion mouse models [3][6][7][8][9][10][11][12][13][14][15] or the administration of nephrotoxic compounds, mainly gentamycin [3][10] or cisplatin [16]. All of these models demonstrated an early increase of miR-21 renal expression [13] that may be prolonged up to 30 days after injury [7]. Following repeated low-intensity injuries, miR-21 increase could thus initially plays a protective role, inducing wound healing and tissue regeneration processes by targeting PTEN [5], PDCD4 [8], PHD2 [15], the MKK3–MAPK–p38 pathway [13], thromospondin-1 [14] and Rab11A [11]. However, there is conflicting evidence regarding the protective or deleterious nature of miR-21. Indeed, in most studies, the inhibition of miR-21 leads to histological damage worsening and decreased renal function [8][10][11] after ischemia reperfusion, even if a protective preconditioning intervention, such as cobalt chloride injection [15], Xenon inhalation [10] or ischemic preconditioning [8], was beforehand applied. By contrast, Chau et al. reported that miR-21 inhibition improves histological injuries and albuminuria seven days after ischemic injury [6]. The main elements that may explain the difference between those studies are the variable miR-21 inhibitor injection schedule and endpoints (in particular euthanasia delay post-injury). We can thus hypothesize that miR-21 plays a protective role at the early stage of ischemia-reperfusion lesions, in particular in preconditioning interventions, but plays a secondary deleterious role once ATN lesions have been initiated.
1.2. Sustained and Persistent Expression of miR-21 Has a Deleterious Impact in Chronic Kidney Diseases
miR-21 has been shown to be elevated in renal tissue, blood or urine in clinical samples from various pathologies. As is consistent with most studies, a miR-21 increase is associated with more severe damages [17][18][19][20][21][22][23][24]. Similarly, an increased expression of miR-21 is unanimously reported in a plethora of chronic kidney disease mouse models, underlining the deleterious role of miR-21 in chronic kidney diseases, including diabetic nephropathy [25][26][27][28][29], unilateral ureteral obstruction [6][25][30] and Alport syndrome [31].
It is noteworthy that most disparate diseases, such as diabetes mellitus, hypertension, Alport Syndrome or acute renal injuries, result in the development of either glomerular or interstitial fibrosis. Converging evidence from computational, biochemical and genetic experiments has indeed shown that miR-21 is a genuine profibrotic miRNA, regardless of the injured organ. In particular, miR-21 is invariably upregulated during the fibrogenic response to tissue injury and promotes the TGF-β signaling pathway, the major driver of tissue fibrosis [32]. In particular, Chau et al. produced a miR-21-null mouse to investigate the role of this miRNA in kidney fibrosis [6]. As is consistent with previous findings reported in cardiac [33] and lung fibrosis [34], injured kidneys from miR-21-deficient mice exhibited less fibrosis. Unexpectedly, the authors further showed that miR-21 primarily regulates the genes involved in lipid metabolism and mitochondrial redox regulation, rather than genes implicated in matrix turnover, inflammation or innate immunity. Indeed, the authors identified PPAR-α, a major transcription factor that regulates a number of lipid oxidation and metabolism pathways, and Mpv17l, which is thought to inhibit ROS formation by mitochondria, as direct targets of miR-21 [6]. This distinct mechanism was explained by the identification of epithelial cells as the major cellular source of increased miR-21 expression. Of particular interest, this study highlights that miR-21 can drive fibrogenesis by several distinct mechanisms, depending on the cellular context (Figure 1).
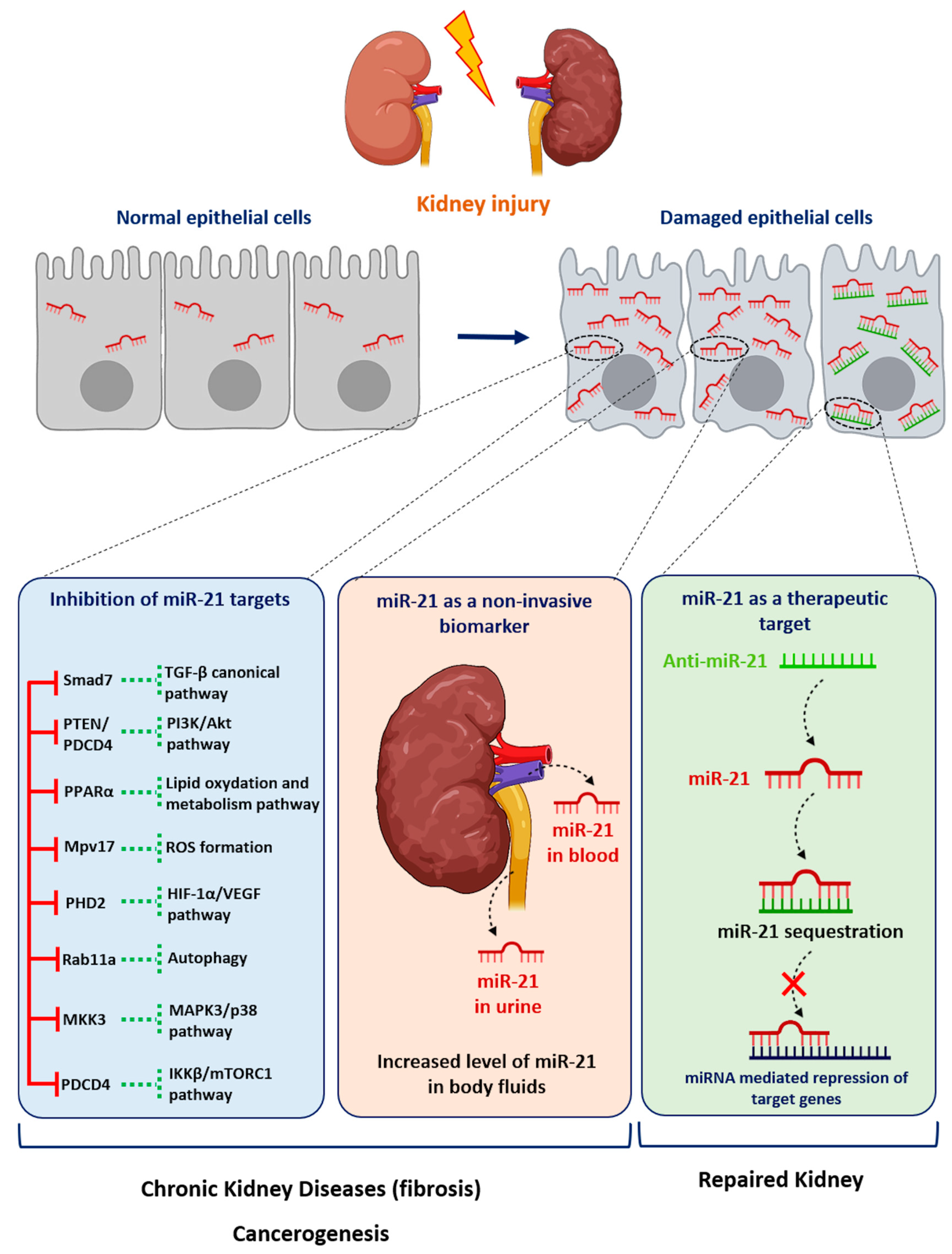
Figure 1. Role of miR-21 in kidney diseases. Following injury, the expression of miR-21 is increased in renal cells. miR-21 promotes kidney diseases by repressing various target genes. Levels of miR-21 can be assessed in urine or blood samples as a biomarker of kidney injury. Given its established pathogenic role in kidney disorders, targeting miR-21 using antisense oligonucleotides may represent a new therapeutic strategy for renal diseases.
2. miR-21 as an “oncomiR”
miRNAs influence numerous cellular processes, including cell cycle regulation, differentiation and apoptosis, and can therefore act as either tumor suppressors or oncogenes [35]. Consequently, alterations in miRNA gene expression have a major impact on tumorigenesis. In particular, the overexpression of miR-21 is associated with many forms of cancer, and functional studies have established this miRNA as a genuine oncomiR (Figure 1). Indeed, many studies have demonstrated that miR-21 has a central role in tumor initiation and progression by targeting critical tumor suppressive genes, such as PTEN or PDCD4. Not surprisingly, comprehensive studies assessing miRNA expression in renal cell carcinoma (RCC) have shown widespread miRNA dysregulation, with many of these aberrantly expressed miRNAs targeting components of key oncogenic networks associated with RCC, including the HIF-, TGF-β- or mTOR-signaling pathways. Several studies on RCC have shown that miR-21 is overexpressed in the clear cell (cRCC) and papillary (pRCC) subtypes of RCC tumors compared with healthy kidneys and benign renal tumors [35][36][37][38][39][40][41]. In cRCC tissue patients, there is no relationship between miR-21 expression and age, laterality or gender [40][41]. Chen et al. have shown, in a cohort of 104 RCC tissue samples, that a higher miR-21 level is associated with larger tumor size, more lymph node metastasis and an advanced TNM stage [41]. In contrast, another study has shown in a cohort of 99 cRCC tissue samples that miR-21 expression was not associated with stage, nuclear Fürhman grade nor patient outcome [40]. Thus, miR-21 expression alone in primary tumors seems of limited interest as a diagnostic or a prognostic biomarker, and should rather be included in miRNA signature [42][43][44][45]. Furthermore, miR-21 is also detected in RCC patient serums, and could be used as biomarker but only in combination with other miRNAs, such as miR-106a, miR-310-3p, miR-150-5p and miR-145-5p [46][47][48]. cRCC accounts for 70–85% of all RCC cases, and is typically highly resistant to conventional therapies [49][50]. Studies from the TCGA uncovered that the altered promoter methylation of miR-21 is associated with aggressive cRCC, suggesting that miR-21 may exert an important oncogenic function in this neoplasia [37][51]. miR-21 is not only upregulated in cRCC but is also involved in cancer progression (proliferation, migration, invasion, epithelial mesenchymal transition) and the cancer stem cell phenotype by targeting tumor suppressor genes such as PTEN, PDCD4, TIMP3 or LATS1 [40][41][52][53][54][55][56][57][58][59][60]. As cRCC is typically highly resistant to conventional systemic therapies [49][50], the identification of new molecular mechanisms driving tumor progression is essential for the rational design of new therapeutic strategies to cure cRCC. In this context, miR-21 has been shown to be involved in the resistance to conventional chemotherapies (paclitaxel, 5-Fluorouracil, topotecan and platinum-based therapy) and targeted therapies such as dovitinib and sorafenib by controlling the expression of genes associated with multi-drug resistance (MDR) and the apoptotic pathway (PTEN, PDCD4) [41][61][62]. Similar to renal fibrosis, miR-21 seems to also be involved in the metabolic shift characterizing renal cancer by targeting PPAR-α, a master regulator of lipid metabolism [63]. miR-21 silencing using antisense oligonucleotide or miR-21 sponge strategies decreases the proliferation, invasion and migration of cRCC cells and also increases the expression of pro-apoptotic markers. Furthermore, the inhibition of miR-21 enhances the sensitivity of cRCC cells to conventional genotoxic drugs, as well as to targeted therapies [40][61][62]. Finally, the inhibition of miR-21 also decreases the expression of MDR genes by a mechanism that remains to be deciphered.
References
- Pickkers, P.; Ostermann, M.; Joannidis, M.; Zarbock, A.; Hoste, E.; Bellomo, R.; Prowle, J.; Darmon, M.; Bonventre, J.V.; Forni, L.; et al. The intensive care medicine agenda on acute kidney injury. Intensive Care Med. 2017, 43, 1198–1209.
- Pavkovic, M.; Robinson-Cohen, C.; Chua, A.S.; Nicoara, O.; Cárdenas-González, M.; Bijol, V.; Ramachandran, K.; Hampson, L.; Pirmohamed, M.; Antoine, D.J.; et al. Detection of Drug-Induced Acute Kidney Injury in Humans Using Urinary KIM-1, miR-21, -200c, and -423. Toxicol. Sci. 2016, 152, 205–213.
- Saikumar, J.; Hoffmann, D.; Kim, T.M.; Gonzalez, V.R.; Zhang, Q.; Goering, P.L.; Brown, R.P.; Bijol, V.; Park, P.J.; Waikar, S.S.; et al. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol. Sci. 2012, 129, 256–267.
- Du, J.; Cao, X.; Zou, L.; Chen, Y.; Guo, J.; Chen, Z.; Hu, S.; Zheng, Z. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS ONE 2013, 8, e63390.
- Ramachandran, K.; Saikumar, J.; Bijol, V.; Koyner, J.L.; Qian, J.; Betensky, R.A.; Waikar, S.S.; Vaidya, V.S. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin. Chem. 2013, 59, 1742–1752.
- Chau, B.N.; Xin, C.; Hartner, J.; Ren, S.; Castano, A.P.; Linn, G.; Li, J.; Tran, P.T.; Kaimal, V.; Huang, X.; et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 2012, 4, 121ra18.
- Godwin, J.G.; Ge, X.; Stephan, K.; Jurisch, A.; Tullius, S.G.; Iacomini, J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc. Natl. Acad. Sci. USA 2010, 107, 14339–14344.
- Xu, X.; Kriegel, A.J.; Liu, Y.; Usa, K.; Mladinov, D.; Liu, H.; Fang, Y.; Ding, X.; Liang, M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012, 82, 1167–1175.
- Kaucsár, T.; Révész, C.; Godó, M.; Krenács, T.; Albert, M.; Szalay, C.I.; Rosivall, L.; Benyó, Z.; Bátkai, S.; Thum, T.; et al. Activation of the miR-17 family and miR-21 during murine kidney ischemia-reperfusion injury. Nucleic Acid Ther. 2013, 23, 344–354.
- Jia, P.; Teng, J.; Zou, J.; Fang, Y.; Zhang, X.; Bosnjak, Z.J.; Liang, M.; Ding, X. miR-21 contributes to xenon-conferred amelioration of renal ischemia-reperfusion injury in mice. Anesthesiology 2013, 119, 621–630.
- Liu, X.; Hong, Q.; Wang, Z.; Yu, Y.; Zou, X.; Xu, L. MiR-21 inhibits autophagy by targeting Rab11a in renal ischemia/reperfusion. Exp. Cell Res. 2015, 338, 64–69.
- Chen, X.; Wang, C.C.; Song, S.M.; Wei, S.Y.; Li, J.S.; Zhao, S.L.; Li, B. The administration of erythropoietin attenuates kidney injury induced by ischemia/reperfusion with increased activation of Wnt/β-catenin signaling. J. Formos. Med. Assoc. 2015, 114, 430–437.
- Li, Z.; Deng, X.; Kang, Z.; Wang, Y.; Xia, T.; Ding, N.; Yin, Y. Elevation of miR-21, through targeting MKK3, may be involved in ischemia pretreatment protection from ischemia-reperfusion induced kidney injury. J. Nephrol. 2016, 29, 27–36.
- Xu, X.; Song, N.; Zhang, X.; Jiao, X.; Hu, J.; Liang, M.; Teng, J.; Ding, X. Renal Protection Mediated by Hypoxia Inducible Factor-1α Depends on Proangiogenesis Function of miR-21 by Targeting Thrombospondin 1. Transplantation 2017, 101, 1811–1819.
- Jiao, X.; Xu, X.; Fang, Y.; Zhang, H.; Liang, M.; Teng, J.; Ding, X. miR-21 contributes to renal protection by targeting prolyl hydroxylase domain protein 2 in delayed ischaemic preconditioning. Nephrology 2017, 22, 366–373.
- Pavkovic, M.; Riefke, B.; Ellinger-Ziegelbauer, H. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology 2014, 324, 147–157.
- Lai, J.Y.; Luo, J.; O’Connor, C.; Jing, X.; Nair, V.; Ju, W.; Randolph, A.; Ben-Dov, I.Z.; Matar, R.N.; Briskin, D.; et al. MicroRNA-21 in glomerular injury. J. Am. Soc. Nephrol. 2015, 26, 805–816.
- Ben-Dov, I.Z.; Muthukumar, T.; Morozov, P.; Mueller, F.B.; Tuschl, T.; Suthanthiran, M. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation 2012, 94, 1086–1094.
- Glowacki, F.; Savary, G.; Gnemmi, V.; Buob, D.; Van der Hauwaert, C.; Lo-Guidice, J.M.; Bouyé, S.; Hazzan, M.; Pottier, N.; Perrais, M.; et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS ONE 2013, 8, e58014.
- Bao, H.; Hu, S.; Zhang, C.; Shi, S.; Qin, W.; Zeng, C.; Zen, K.; Liu, Z. Inhibition of miRNA-21 prevents fibrogenic activation in podocytes and tubular cells in IgA nephropathy. Biochem. Biophys. Res. Commun. 2014, 444, 455–460.
- McClelland, A.D.; Herman-Edelstein, M.; Komers, R.; Jha, J.C.; Winbanks, C.E.; Hagiwara, S.; Gregorevic, P.; Kantharidis, P.; Cooper, M.E. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin. Sci. 2015, 129, 1237–1249.
- Pezzolesi, M.G.; Satake, E.; McDonnell, K.P.; Major, M.; Smiles, A.M.; Krolewski, A.S. Circulating TGF-β1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes 2015, 64, 3285–3293.
- Zununi Vahed, S.; Omidi, Y.; Ardalan, M.; Samadi, N. Dysregulation of urinary miR-21 and miR-200b associated with interstitial fibrosis and tubular atrophy (IFTA) in renal transplant recipients. Clin. Biochem. 2017, 50, 32–39.
- Hennino, M.F.; Buob, D.; Van der Hauwaert, C.; Gnemmi, V.; Jomaa, Z.; Pottier, N.; Savary, G.; Drumez, E.; Noël, C.; Cauffiez, C.; et al. miR-21-5p renal expression is associated with fibrosis and renal survival in patients with IgA nephropathy. Sci. Rep. 2016, 6, 27209.
- Zhong, X.; Chung, A.C.; Chen, H.Y.; Meng, X.M.; Lan, H.Y. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J. Am. Soc. Nephrol. 2011, 22, 1668–1681.
- Wang, J.; Gao, Y.; Ma, M.; Li, M.; Zou, D.; Yang, J.; Zhu, Z.; Zhao, X. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem. Biophys. 2013, 67, 537–546.
- Wang, J.Y.; Gao, Y.B.; Zhang, N.; Zou, D.W.; Wang, P.; Zhu, Z.Y.; Li, J.Y.; Zhou, S.N.; Wang, S.C.; Wang, Y.Y.; et al. miR-21 overexpression enhances TGF-β1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol. Cell. Endocrinol. 2014, 392, 163–172.
- Wang, J.; Duan, L.; Tian, L.; Liu, J.; Wang, S.; Gao, Y.; Yang, J. Serum miR-21 may be a Potential Diagnostic Biomarker for Diabetic Nephropathy. Exp. Clin. Endocrinol. Diabetes 2016, 124, 417–423.
- Wu, H.; Kong, L.; Tan, Y.; Epstein, P.N.; Zeng, J.; Gu, J.; Liang, G.; Kong, M.; Chen, X.; Miao, L.; et al. C66 ameliorates diabetic nephropathy in mice by both upregulating NRF2 function via increase in miR-200a and inhibiting miR-21. Diabetologia 2016, 59, 1558–1568.
- Zarjou, A.; Yang, S.; Abraham, E.; Agarwal, A.; Liu, G. Identification of a microRNA signature in renal fibrosis: Role of miR-21. Am. J. Physiol. Ren. Physiol. 2011, 301, F793–F801.
- Gomez, I.G.; MacKenna, D.A.; Johnson, B.G.; Kaimal, V.; Roach, A.M.; Ren, S.; Nakagawa, N.; Xin, C.; Newitt, R.; Pandya, S.; et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Investig. 2015, 125, 141–156.
- Huang, Y.; He, Y.; Li, J. MicroRNA-21: A central regulator of fibrotic diseases via various targets. Curr. Pharm. Des. 2015, 21, 2236–2242.
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984.
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597.
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234.
- Faragalla, H.; Youssef, Y.M.; Scorilas, A.; Khalil, B.; White, N.M.; Mejia-Guerrero, S.; Khella, H.; Jewett, M.A.; Evans, A.; Lichner, Z.; et al. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J. Mol. Diagn. 2012, 14, 385–392.
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209.
- Kowalczyk, A.E.; Krazinski, B.E.; Godlewski, J.; Grzegrzolka, J.; Kiewisz, J.; Kwiatkowski, P.; Sliwinska-Jewsiewicka, A.; Dziegiel, P.; Kmiec, Z. SATB1 is Down-regulated in Clear Cell Renal Cell Carcinoma and Correlates with miR-21-5p Overexpression and Poor Prognosis. Cancer Genom. Proteom. 2016, 13, 209–217.
- Petrozza, V.; Carbone, A.; Bellissimo, T.; Porta, N.; Palleschi, G.; Pastore, A.L.; Di Carlo, A.; Della Rocca, C.; Fazi, F. Oncogenic MicroRNAs Characterization in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 29219–29225.
- Gaudelot, K.; Gibier, J.B.; Pottier, N.; Hémon, B.; Van Seuningen, I.; Glowacki, F.; Leroy, X.; Cauffiez, C.; Gnemmi, V.; Aubert, S.; et al. Targeting miR-21 decreases expression of multi-drug resistant genes and promotes chemosensitivity of renal carcinoma. Tumor Biol. 2017, 39, 1010428317707372.
- Chen, J.; Gu, Y.; Shen, W. MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4566–4576.
- Tang, K.; Xu, H. Prognostic value of meta-signature miRNAs in renal cell carcinoma: An integrated miRNA expression profiling analysis. Sci. Rep. 2015, 5, 10272.
- Huang, M.; Zhang, T.; Yao, Z.Y.; Xing, C.; Wu, Q.; Liu, Y.W.; Xing, X.L. MicroRNA related prognosis biomarkers from high throughput sequencing data of kidney renal clear cell carcinoma. BMC Med. Genom. 2021, 14, 72.
- Guo, Y.; Li, X.; Zheng, J.; Fang, J.; Pan, G.; Chen, Z. Identification of a novel immune-related microRNA prognostic model in clear cell renal cell carcinoma. Transl. Androl. Urol. 2021, 10, 888–899.
- Zhao, Y.; Tao, Z.; Chen, X. Identification of the miRNA-mRNA regulatory pathways and a miR-21-5p based nomogram model in clear cell renal cell carcinoma. PeerJ 2020, 8, e10292.
- Tusong, H.; Maolakuerban, N.; Guan, J.; Rexiati, M.; Wang, W.G.; Azhati, B.; Nuerrula, Y.; Wang, Y.J. Functional analysis of serum microRNAs miR-21 and miR-106a in renal cell carcinoma. Cancer Biomark. 2017, 18, 79–85.
- Kalogirou, C.; Ellinger, J.; Kristiansen, G.; Hatzichristodoulou, G.; Kübler, H.; Kneitz, B.; Busch, J.; Fendler, A. Identification of miR-21-5p and miR-210-3p serum levels as biomarkers for patients with papillary renal cell carcinoma: A multicenter analysis. Transl. Androl. Urol. 2020, 9, 1314–1322.
- Chen, X.; Li, R.; Li, X.; Peng, X.; Zhang, C.; Liu, K.; Huang, G.; Lai, Y. Identification of a four-microRNA panel in serum for screening renal cell carcinoma. Pathol. Res. Pract. 2021, 227, 153625.
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009.
- Wettersten, H.I.; Aboud, O.A.; Lara, P.N., Jr.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419.
- Lu, J.; Tan, T.; Zhu, L.; Dong, H.; Xian, R. Hypomethylation Causes MIR21 Overexpression in Tumors. Mol. Ther. Oncolytics 2020, 18, 47–57.
- Wu, T.K.; Wei, C.W.; Pan, Y.R.; Hsu, R.J.; Wu, C.Y.; Yu, Y.L. The uremic toxin p-cresyl sulfate induces proliferation and migration of clear cell renal cell carcinoma via microRNA-21/ HIF-1α axis signals. Sci. Rep. 2019, 9, 3207.
- An, F.; Liu, Y.; Hu, Y. miR-21 inhibition of LATS1 promotes proliferation and metastasis of renal cancer cells and tumor stem cell phenotype. Oncol. Lett. 2017, 14, 4684–4688.
- Yuan, H.; Xin, S.; Huang, Y.; Bao, Y.; Jiang, H.; Zhou, L.; Ren, X.; Li, L.; Wang, Q.; Zhang, J. Downregulation of PDCD4 by miR-21 suppresses tumor transformation and proliferation in a nude mouse renal cancer model. Oncol. Lett. 2017, 14, 3371–3378.
- Cao, J.; Liu, J.; Xu, R.; Zhu, X.; Liu, L.; Zhao, X. MicroRNA-21 stimulates epithelial-to-mesenchymal transition and tumorigenesis in clear cell renal cells. Mol. Med. Rep. 2016, 13, 75–82.
- Li, X.; Xin, S.; He, Z.; Che, X.; Wang, J.; Xiao, X.; Chen, J.; Song, X. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell. Physiol. Biochem. 2014, 33, 1631–1642.
- Bera, A.; Ghosh-Choudhury, N.; Dey, N.; Das, F.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. NFκB-mediated cyclin D1 expression by microRNA-21 influences renal cancer cell proliferation. Cell. Signal. 2013, 25, 2575–2586.
- Bera, A.; Das, F.; Ghosh-Choudhury, N.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKβ-mTORC1 axis to regulate renal cancer cell invasion. Exp. Cell Res. 2014, 328, 99–117.
- Dey, N.; Das, F.; Ghosh-Choudhury, N.; Mandal, C.C.; Parekh, D.J.; Block, K.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE 2012, 7, e37366.
- Zhang, H.; Guo, Y.; Shang, C.; Song, Y.; Wu, B. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology 2012, 80, 1298–1302.e1.
- Naro, Y.; Ankenbruck, N.; Thomas, M.; Tivon, Y.; Connelly, C.M.; Gardner, L.; Deiters, A. Small Molecule Inhibition of MicroRNA miR-21 Rescues Chemosensitivity of Renal-Cell Carcinoma to Topotecan. J. Med. Chem. 2018, 61, 5900–5909.
- Liu, L.; Pang, X.; Shang, W.; Xie, H.; Feng, Y.; Feng, G. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle 2019, 18, 257–263.
- Goujon, M.; Woszczyk, J.; Gaudelot, K.; Swierczewski, T.; Fellah, S.; Gibier, J.B.; Van Seuningen, I.; Larrue, R.; Cauffiez, C.; Gnemmi, V.; et al. A Double-Negative Feedback Interaction between miR-21 and PPAR-α in Clear Renal Cell Carcinoma. Cancers 2022, 14, 795.
More
