Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Nando Dulal Das.
The physical interactions between enhancers and promoters create chromatin conformations involved in gene regulation. Although it is not entirely comprehensive how chromatin-mediated enhancer–promoter (E–P) interactions with various histone marks can affect gene expression, this proximity has been observed in multiple systems at multiple loci and is thought to be essential to control gene expression.
- gene expression
- enhancer
- promoter
- histone modifications
1. Introduction
The spatial organization of the genome into transcriptionally active and silenced chromatin plays a fundamental role in the three-dimensional (3D) architecture required for the regulation of eukaryotic genes [1,2][1][2]. Genomic sequences are partitioned into one of two nuclear compartments called the A/B compartments: the A compartment is an ‘open’ state that allows for the transcription of the associated genes (euchromatin), and the B compartment is a ‘closed’ state associated with inactive genes (heterochromatin) [3]. The compacted chromatin of heterochromatin is assumed to be inaccessible to transcriptional machinery and resistant to chromatin remodeling; this condensed state is thus accepted as a major hallmark of repressed chromatin, which comprises silenced genes [4]. Heterochromatin is further divided into two types, constitutive and facultative. In constitutive heterochromatin, repetitive sequences such as pericentromeric regions are organized into silent nuclear compartments that are highly enriched in trimethylated histone H3 lysine 9 (H3K9me3) and methylated DNA [5]. In contrast, facultative heterochromatin consists of transcriptionally silent regions that become activated depending on the context [6].
In one key mechanism, gene transcription is regulated through chromatin loops that form between promoters and various regulatory elements, including enhancers [3,7][3][7]. Enhancers are cis-elements that contain diverse DNA sequences to which various transcription factors (TFs) and transcriptional co-activators bind and that are enriched with various histone modifications that facilitate gene transcription [8,9,10][8][9][10].
2. Histone Modifications Involved in E–P Interactions
Histone modification is widely used as a means to classify enhancers according to their activity: H3K4me1 and the binding of trithorax-related mixed lineage leukemia (MLL) complex define primed or active enhancers; H3K27me3 is a key marker of poised or inactive enhancers; histone H3 lysine 27 acetylation (H3K27ac) is a hallmark of transcriptionally active enhancers (Figure 1) [11,12,13,14][11][12][13][14]. Recent trends highlight that rather than defining active enhancers with H3K27ac, different histone acetylation marks, such as simultaneous acetylation of histone H4 at both K5 and K8 (H4K5acK8ac) [15], Das et al., [submitted], histone H2B N-terminus multisite lysine (e.g., K5, K12, K16, and K20) acetylation (H2BNTac [16]), H3K122ac [17], and H4K16ac [18], to define active enhancers are emerging. A large number of histone modifications have been implicated in gene transcription, where H3K4me3 has been associated with gene promoter regions [19]. Table 1 summarizes the most well-characterized histone modifications and their involvement in E–P interactions.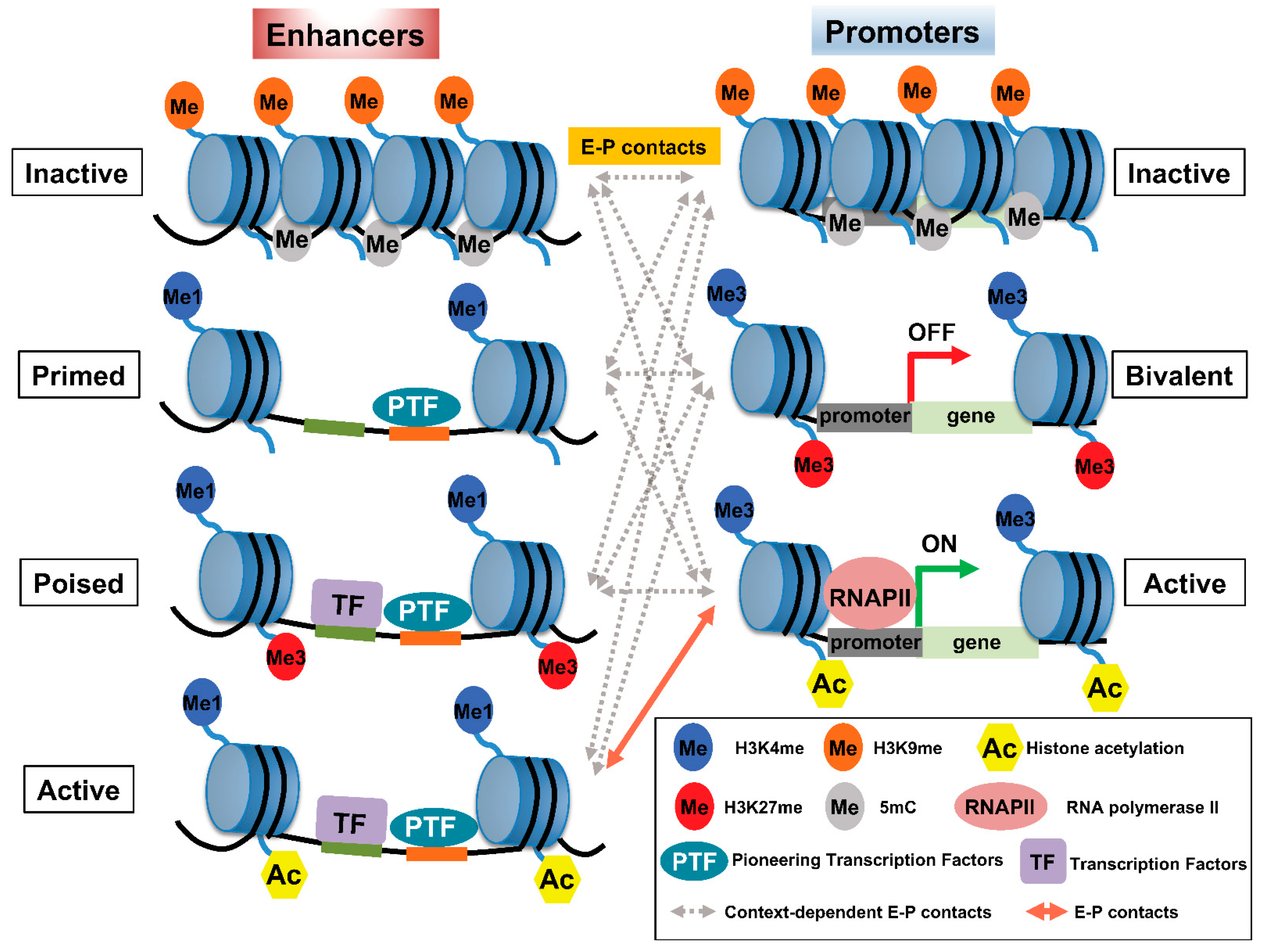
Figure 1. Pattern of histone modifications defines enhancer–promoter (E–P) states. (Left panel) The pattern of histone modifications in various enhancer states. Inactive enhancers are characterized by high nucleosome density and enrichment of H3K9me2/3 and DNA methylation. At primed enhancers, pioneering TFs (PTF) bind to their target sites and recruit MLL3/4 complexes to decorate H3K4me1. PTF binding is accompanied by nucleosome remodeling. H3K27me3 may contribute to the poised status of an enhancer to prevent premature activation. In addition, poised enhancers may allow lineage-specific TFs (LTF) to bind and recruit histone acetyltransferases, such as p300/CBP, and H3K27 demethylase(s) to prepare the enhancer for rapid activation. Active enhancers contain not only H3K27ac—the hallmark of an active enhancer—but also H4K5acK8ac, H2BNTac, and potentially acetylation at various residues of other histones. (Right panel) The pattern of histone modifications in various promoter states. Promoter activity is highly correlated with enhancer activity and the associated epigenetic landscape. Inactive promoters carry high levels of H3K9me2/3 and DNA methylation. The vast majority of each promoter is marked by H3K4me3, whereas other parts of the promoters are marked by H3K27me3, thus constituting so-called ‘bivalent promoters,’ which are repressed by polycomb group complexes. In comparison, active promoters gain histone acetylation marks. The epigenetic landscape of enhancers and promoters can be variably altered in response to external signals. However, the timing of and how E–P interactions facilitate gene expression is not well understood and may involve context-dependent interactions among enhancers and promoters at various states.
Table 1.
Summary of histone modifications and their putative role in enhancer–promoter (E–P) interactions and transcription.
| Histone Modification | Putative Role in E–P Interactions and Transcription | Most Enriched Region | Reference |
|---|---|---|---|
| H3K4me3 | Activation | Promoters, bivalent promoters | [19] |
| H3K4me1 | Activation | Enhancers | [14] |
| H3K27ac | Activation | Enhancers, promoters | [11,12,13,14][11][12][13][14] |
| H4K5acK8ac | Activation | Enhancers, promoters | [15] |
| H4K16ac | Activation | Enhancers | [18] |
| H2BNTac (K5, K12, K16, K20) | Activation | Enhancers | [16] |
| H3K27me3 | Repression | Bivalent Promoters, poised enhancers | [30,31,32,33,31][32][33]34][30][[34] |
References
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43.
- Yu, M.; Ren, B. The Three-Dimensional Organization of Mammalian Genomes. Annu. Rev. Cell Dev. Biol. 2017, 33, 265–289.
- Pombo, A.; Dillon, N. Three-dimensional genome architecture: Players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015, 16, 245–257.
- Workman, J.L.; Kingston, R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998, 67, 545–579.
- Padeken, J.; Methot, S.P.; Gasser, S.M. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat. Rev. Mol. Cell Biol. 2022, 23, 623–640.
- Trojer, P.; Reinberg, D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol. Cell 2007, 28, 1–13.
- Schoenfelder, S.; Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455.
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112.
- Kim, S.; Shendure, J. Mechanisms of Interplay between Transcription Factors and the 3D Genome. Mol. Cell 2019, 76, 306–319.
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626.
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837.
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936.
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318.
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011, 470, 279–283.
- Handoko, L.; Kaczkowski, B.; Hon, C.C.; Lizio, M.; Wakamori, M.; Matsuda, T.; Ito, T.; Jeyamohan, P.; Sato, Y.; Sakamoto, K.; et al. JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states. Epigenetics 2018, 13, 410–431.
- Narita, T.; Higashijima, Y.; Kilic, S.; Liebner, T.; Walter, J.; Choudhary, C. A unique H2B acetylation signature marks active enhancers and predicts their target genes. bioRxiv 2022.
- Pradeepa, M.M.; Grimes, G.R.; Kumar, Y.; Olley, G.; Taylor, G.C.; Schneider, R.; Bickmore, W.A. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 2016, 48, 681–686.
- Taylor, G.C.; Eskeland, R.; Hekimoglu-Balkan, B.; Pradeepa, M.M.; Bickmore, W.A. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 2013, 23, 2053–2065.
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080.
- Goudarzi, A.; Shiota, H.; Rousseaux, S.; Khochbin, S. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J. Mol. Biol. 2014, 426, 3342–3349.
- Shiota, H.; Barral, S.; Buchou, T.; Tan, M.; Coute, Y.; Charbonnier, G.; Reynoird, N.; Boussouar, F.; Gerard, M.; Zhu, M.; et al. Nut Directs p300-Dependent, Genome-Wide H4 Hyperacetylation in Male Germ Cells. Cell Rep. 2018, 24, 3477–3487.e6.
- Alekseyenko, A.A.; Walsh, E.M.; Wang, X.; Grayson, A.R.; Hsi, P.T.; Kharchenko, P.V.; Kuroda, M.I.; French, C.A. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 2015, 29, 1507–1523.
- Baud, M.G.J.; Lin-Shiao, E.; Cardote, T.; Tallant, C.; Pschibul, A.; Chan, K.H.; Zengerle, M.; Garcia, J.R.; Kwan, T.T.; Ferguson, F.M.; et al. Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science 2014, 346, 638–641.
- Dey, A.; Chitsaz, F.; Abbasi, A.; Misteli, T.; Ozato, K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 2003, 100, 8758–8763.
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Muller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231.
- Devaiah, B.N.; Case-Borden, C.; Gegonne, A.; Hsu, C.H.; Chen, Q.; Meerzaman, D.; Dey, A.; Ozato, K.; Singer, D.S. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016, 23, 540–548.
- Wang, R.; Li, Q.; Helfer, C.M.; Jiao, J.; You, J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J. Biol. Chem. 2012, 287, 10738–10752.
- Eagen, K.P.; French, C.A. Supercharging BRD4 with NUT in carcinoma. Oncogene 2021, 40, 1396–1408.
- Rosencrance, C.D.; Ammouri, H.N.; Yu, Q.; Ge, T.; Rendleman, E.J.; Marshall, S.A.; Eagen, K.P. Chromatin Hyperacetylation Impacts Chromosome Folding by Forming a Nuclear Subcompartment. Mol. Cell 2020, 78, 112–126.e12.
- Sharif, J.; Endo, T.A.; Ito, S.; Ohara, O.; Koseki, H. Embracing change to remain the same: Conservation of polycomb functions despite divergence of binding motifs among species. Curr. Opin. Cell Biol. 2013, 25, 305–313.
- Voigt, P.; Tee, W.W.; Reinberg, D. A double take on bivalent promoters. Genes Dev. 2013, 27, 1318–1338.
- Ferrari, K.J.; Scelfo, A.; Jammula, S.; Cuomo, A.; Barozzi, I.; Stutzer, A.; Fischle, W.; Bonaldi, T.; Pasini, D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 2014, 53, 49–62.
- Ngan, C.Y.; Wong, C.H.; Tjong, H.; Wang, W.; Goldfeder, R.L.; Choi, C.; He, H.; Gong, L.; Lin, J.; Urban, B.; et al. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat. Genet. 2020, 52, 264–272.
- Zeitlinger, J.; Stark, A.; Kellis, M.; Hong, J.W.; Nechaev, S.; Adelman, K.; Levine, M.; Young, R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007, 39, 1512–1516.
- Spivakov, M.; Fisher, A.G. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007, 8, 263–271.
- Hafner, A.; Boettiger, A. The spatial organization of transcriptional control. Nat. Rev. Genet. 2022.
- Robson, M.I.; Ringel, A.R.; Mundlos, S. Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol. Cell 2019, 74, 1110–1122.
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C.; Heckert, A.; Banala, S.; Lavis, L.; Darzacq, X.; et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361, eaar2555.
- Zabidi, M.A.; Arnold, C.D.; Schernhuber, K.; Pagani, M.; Rath, M.; Frank, O.; Stark, A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 2015, 518, 556–559.
- Cai, Y.; Zhang, Y.; Loh, Y.P.; Tng, J.Q.; Lim, M.C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat. Commun. 2021, 12, 719.
- Sungalee, S.; Liu, Y.; Lambuta, R.A.; Katanayeva, N.; Donaldson Collier, M.; Tavernari, D.; Roulland, S.; Ciriello, G.; Oricchio, E. Histone acetylation dynamics modulates chromatin conformation and allele-specific interactions at oncogenic loci. Nat. Genet. 2021, 53, 650–662.
More
