Colorectal cancer (CRC) represents the third most prevalent cancer worldwide and a leading cause of mortality among the population of western countries. However, CRC is frequently a preventable malignancy due to various screening tests being available. While failing to obtain real-time data, current screening methods (either endoscopic or stool-based tests) also require disagreeable preparation protocols and tissue sampling through invasive procedures, rendering adherence to CRC screening programs suboptimal. In this context, the necessity for novel, less invasive biomarkers able to identify and assess cancer at an early stage is evident. Liquid biopsy comes as a promising minimally invasive diagnostic tool, able to provide comprehensive information on tumor heterogeneity and dynamics during carcinogenesis.
- liquid biopsy
- circulating tumor cells
- circulating nucleic acids
- circulating DNA
- microRNA
- exosomes
- colorectal cancer
- screening
1. Introduction
2. Liquid Biopsy
Recent research has been shedding light on a new diagnostic approach suited for cancer patients, the liquid biopsy. Liquid biopsy comes as a simple, minimally invasive diagnostic tool, attempting to overcome the limitations of conventional tissue biopsy by providing more comprehensive data on tumor heterogeneity and dynamics at different junctures in cancer development [13]. Liquid biopsy refers to the biological fluids obtained from cancer patients and submitted to extensive analysis in order to isolate biomarkers indicative of malignancy. The liquid samples considered for testing can include any biological fluid (e.g., urine, pleural effusion, ascites, sputum, or cerebrospinal fluid), however, the main focus is peripheral blood [14]. The main components of liquid biopsies are circulating tumor cells (CTCs), circulating nucleic acids (circulating tumor DNA and circulating microRNAs) and extracellular vesicles (exosomes and microvesicles) [12,15][12][15] (Figure 1). Liquid biopsies allow a comprehensive analysis of plasma, also considered the somatic component of the blood, which can be manipulated for the isolation of CTCs, circulating nucleic acids and exosomes [16]. Somatic mutations can be detected through a thorough examination of these plasma components, which are shed into the bloodstream directly from the primary tumor and distant metastasis, therefore offering an extensive characterization of the tumor mass [17]. In this context, liquid biopsy analytes are currently finding their clinical application in the setting of CRC screening.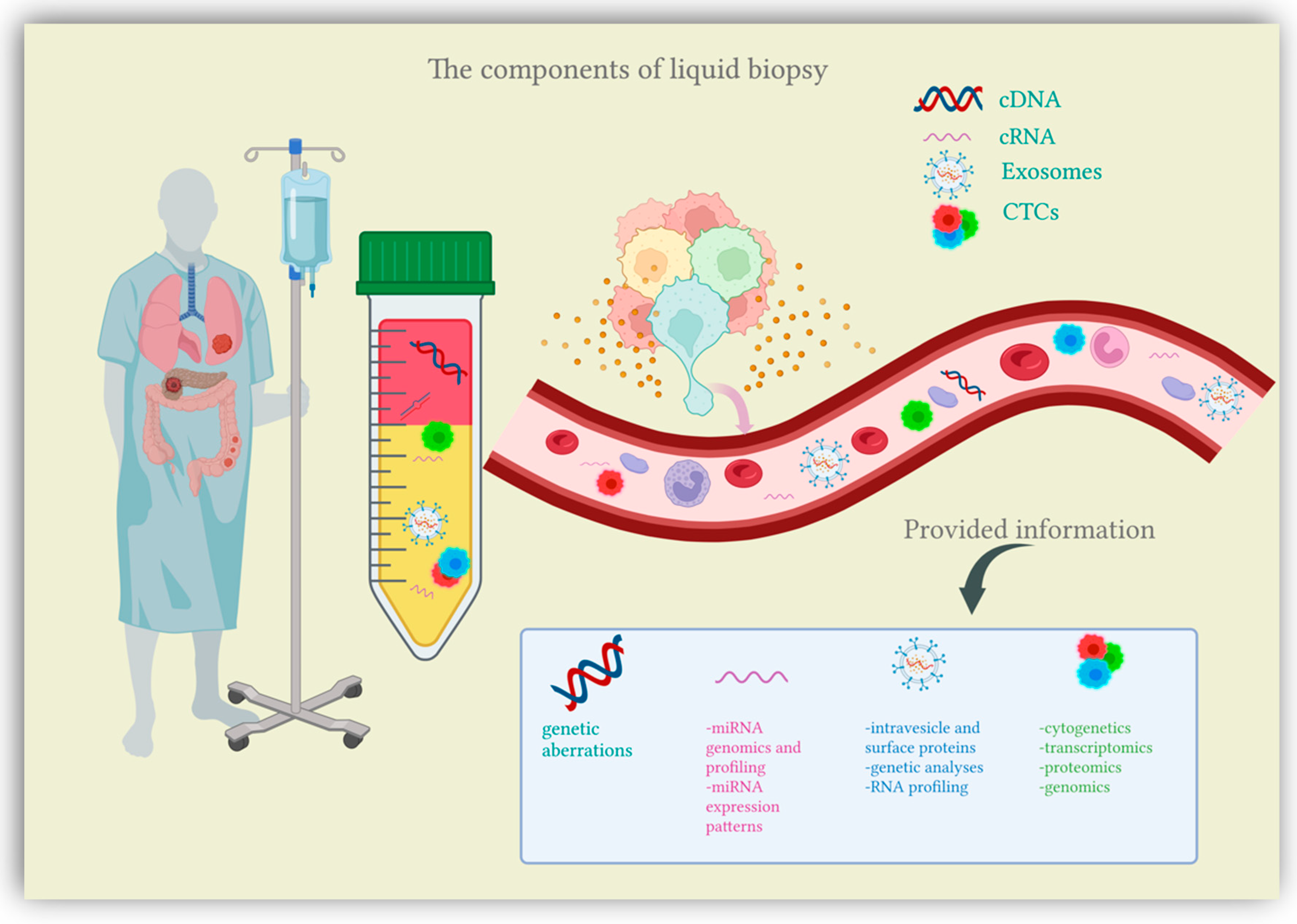
2.1. Circulating Tumor Cells (CTCs)
Originally described in 1869, CTCs are now gaining clinical importance in the management of patients with cancer [18]. CTCs define cells derived from the primary tumor, metastases and recurrence sites that enter the circulatory system either as individual cells or as clusters [19]. Tumor cells constitute these clusters alone or in association with fibroblasts, leukocytes, endothelial cells and platelets, forming tumor microemboli more resistant against the aggression of the host’s immune system [20]. Once they have entered the bloodstream, CTCs bear the capacity to seed the disease to secondary sites, causing tumor metastases in distant organs and disease relapses [19] (Figure 2). Furthermore, CTCs have shown great plasticity through their ability to undergo epithelial-to-mesenchymal transition (EMT) [21]. As most CTCs entering the bloodstream are exposed to mechanical and environmental factors (oxidative stress, shear stress, immunological response, and the absence of growth factors), their clearance is particularly rapid, with a half-life usually limited to 1–2 h [19,22][19][22]. The number of CTCs varies between 1 to 10 cells per 10 mL of blood, with higher counts detected in metastatic patients compared to early-stage cancers [23,24][23][24]. Given the extremely low count of CTCs, their adequate quantification requires special enrichment, detection and characterization technologies.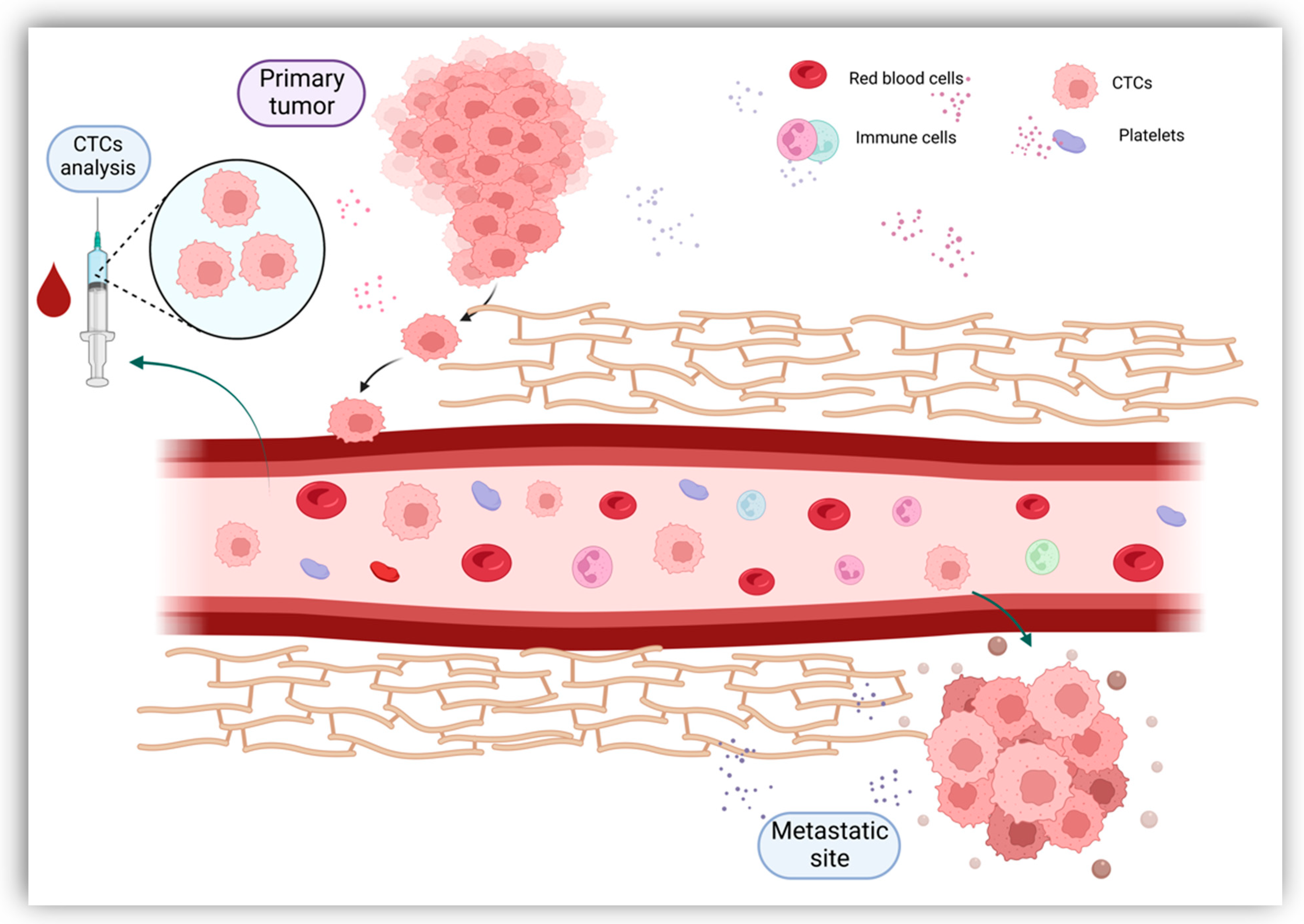
2.2. Circulating Nucleic Acids (CNAs)
The analysis of circulating nucleic acids (CNAs) represents a novel minimally invasive approach, able to assess the tumor and its molecular characteristics while allowing a more accurate description of tumor heterogeneity and evolution in time. CNAs, more specifically, circulating tumor DNA (ctDNA) and circulating microRNA (miRNA), are generally isolated from blood; however, other biological fluids could represent a source of CNAs (saliva, cerebrospinal fluid, pleural effusions, urine and stool, etc.) [53]. CNAs enter the bloodstream by passive release or through active secretion. The passive release of CNAs results from increased production and subsequent shedding of cell debris produced by tumor necrosis and apoptosis [12,54,55][12][54][55]. The active release mechanism sees CNAs packed inside extracellular vesicles such as exosomes to be further secreted by tumor cells [56].2.3. Exosomes
Present in all biological fluids, exosomes represent cell-derived nanovesicles with sizes ranging from 30 to 150 nm in diameter [114][57]. Exosomes develop from the intracellular endosomal compartment following a process of inward expansion from the limiting membrane that generates multivesicular bodies (MVBs) [115][58]. MVBs are then discharged into the extracellular matrix as a result of their fusion with the cytoplasmic membrane, releasing their content in the form of exosomes [116][59]. Exosomal secretion occurs in both physiological and pathological processes, with various types of cells producing exosomes (cancer cells, as well as adipocytes, immune cells and brain cells) [117][60]. Exosomes play a series of roles within the cell, notably the removal of waste, antigen presentation and cytokine release [118][61]. However, their essential function is intercell communication of molecular information [119][62]. Exosomal cargo, mainly cytoplasmic components such as proteins, bio-functional lipids and nucleic acids (mRNA, miRNA and DNA fragments) are key players in the signaling pathways between cells [120][63]. Communication between exosomes and their target cells can be acquired through interaction with surface-expressed ligands, through phagocytosis, or by exosomal fusion with cell membranes [121][64]. An increasing amount of evidence has identified exosomes as essential participants in cancer development processes. Exosomes secreted by tumor cells have been found responsible for alterations in the immune response that lead to suppression of antitumor response [122][65]. In addition, exosomes play important roles in tumor microenvironment (TME) remodeling, angiogenesis and tumor growth, therefore favoring disease progression [118][61]. Exosomes were found to promote EMT, migration and invasion, causing distant cancer dissemination through various proteins and miRNAs [123][66]. Furthermore, research has proved that exosomal signaling pathways are also involved in therapy resistance [123,124][66][67]. Since exosomes have the ability to target specific cells [125][68], technologies have investigated the possibility of manipulating exosomes for therapeutical purposes, by using them as potential vehicles for drug delivery inside tumor cells [126][69]. Exosome concentration in biological fluids is relatively low, thus making their isolation, detection and further analysis relatively challenging. Isolation of exosomes can be achieved based on their physical properties (size and density), electromagnetic characteristics, or according to their immunological properties [127][70]. Isolation through ultracentrifugation represents the gold standard, however, other techniques, such as ultrafiltration, chromatography, hydrostatic filtration dialysis, precipitation, microfluidic chips, or immunoaffinity-based methods, may also facilitate exosome isolation [127][70]. Following isolation, the detection and characterization of exosomes from a morphological point of view is obtained through transmission electron microscopy [128][71]. Nanoparticle tracking analysis can determine size and concentration characteristics [129][72]. Protein expression and functions can be assessed using Western blot and ELISA methods, while exosome content can be identified via spectrophotometric assays and different other focused approaches (e.g., RT-qPCR, Western blot and mass spectrometry) [127,130][70][73]. Exosomal microRNAs have been attracting considerable attention as promising biomarkers suitable for cancer diagnosis. Multiple exosomal miRNAs have been studied, with some of them showing substantial value in identifying early cases of CRC . Research has identified several significantly up-regulated miRNAs in the setting of CRC, including cases of early-stage disease. One study found miRNA-23a to have an important diagnostic accuracy with an area under the curve (AUC) of 0.953, while miRNA-1246 and miRNA-21 also showed encouraging results [131][74]. Overexpression of exosomal miRNA-23a was successfully confirmed by another study that additionally investigated the diagnostic value of exosomal miRNA-301a [132][75]. Another study conducted by a Chinese team demonstrated the overexpression of exosomal miRNA-6803-5p in the serum of CRC patients [133][76]. Similarly, the expression of miRNA-486-5p in plasma exosomes was found to be significantly upregulated in CRC patients, corresponding to disease staging [134][77]. Circulating exosomal miRNA-125a-3p is another upregulated analyte with confirmed value in detecting early-stage CRC patients. The predictive accuracy was improved when associating miRNA-125a-3p with CEA analysis [135][78]. The same study found exosomal miRNA-320c to be up-regulated in plasma samples collected from CRC patients, validating it as a potential biomarker for early diagnosis [135][78]. Downregulation of serum exosomal miRNA-150-5p proved to be a conclusive indicator of colorectal malignancy, while combined analysis with CEA resulted in a higher diagnostic value for CRC identification [136][79]. Interestingly, expression of exosomal miRNA-92b was found to be significantly reduced in patients presenting with CRC, as well as individuals with adenomas of the colon [137][80]. Furthermore, serum samples and serum exosomes isolated from CRC patients also showed a considerable expression of mRNA-196b-5p [138][81]. Additional data identified a downregulation in exosomal miRNA-139-3p expression in plasma samples collected from patients with CRC, correlating with disease aggressiveness [139][82]. Furthermore, plasma collected from CRC patients was found to express important levels of exosomal miRNA-27a and miRNA-130a [140][83]. A different study found upregulation of miRNA-1359 in exosomes isolated from CRC patients, leading to an accurate differentiation of CRC cases from healthy individuals [141][84]. Exosomal miRNAs are receiving a growing interest, with several other studies aiming to identify and validate promising biomarkers for early CRC diagnosis and population screening [131,142,143,144][74][85][86][87]. In the given circumstances, the need to improve the accuracy of these biomarkers is evident.3. The Future Applications of Liquid Biopsies in CRC
Over the past few years, immunotherapy has changed the treatment paradigm for many cancer types. In CRC, the MSI-high phenotype was associated with a significant response to immune checkpoint inhibitors (ICIs) [145][88]. Moreover, the tumor mutational burden (TMB), referring to the number of somatic mutations, was significantly correlated with the outcome of CRC patients treated with ICIs [146][89]. Despite the promising results reported by the scientific community, we face a poor prediction of response to ICIs, along with important rates of innate or acquired resistance leading to heterogenous responses among patients [147][90]. Biomarker-directed use immunotherapy is an important frontier in precision medicine. To date, liquid biopsies are investigated for use as biomarkers to predict and evaluate the response to immunotherapy. CTCs, circulating DNA (cDNA), circulating RNA (cRNA) and exosomes hold a generous amount of tumor-related information. Moreover, liquid biopsies may provide a more comprehensive and dynamic overview of the tumor microenvironment and heterogeneity than single-site tissue biopsies [148][91]. The utility of cDNA as a prognostic and predictive biomarker for immunotherapy was shown in a phase II trial including patients with advanced or metastatic solid tumors treated with an anti-PD1 agent. The study reported that higher pretreatment variant allele frequencies (VAF) were associated with a poorer OS. However, on-treatment VAF and on-treatment reduction in VAF were correlated with longer PFS and OS [149][92]. These findings suggest that on-treatment cDNA variations can predict a beneficial response to ICIs. Similarly, another phase II prospective trial assessed cDNA in patients with advanced solid tumors under treatment with pembrolizumab. Low baseline cDNA levels were correlated with PFS, OS, clinical response and clinical benefit. Moreover, the reduction of cDNA after only two cycles of pembrolizumab and cDNA clearance on-treatment identified a good prognosis subset of patients [150][93]. A better selection of the patients receiving immunotherapy could be guided by specific somatic mutations detectable in cDNA. In this regard, a genomic mutation signature was developed to characterize immunophenotypes and predict response to immunotherapy in gastrointestinal cancers [151][94]. As mentioned above, the number of somatic mutations known as TMB represents an independent predictor of response to ICIs in many solid tumors, including CRC. TMB-high cases (≥20 mutations/megabase) typically occur in microsatellite instable tumors (MSI) or those harboring pathogenic mutations emerging in the DNA polymerases POLD and POLE [152][95]. Currently, the standard evaluation for TMB is based on tissue samples and encounters many limitations. Tissue-based biopsies cannot correctly assess intratumoral heterogeneity or evaluate the changes occurring during treatment [153][96]. In this regard, cDNA-based evaluation of TMB (cTMB) is currently being investigated in clinical trials, with encouraging results obtained in non-small cell lung cancer (NSCLC). The concordance between tissue-based TMB and cDNA-based TMB was strong in clinical trials, suggesting that cTMB could be a feasible predictive biomarker for ICIs [154][97]. Due to recent technological advances, circulating tumor cell PD-L1 expression is being investigated in clinical trials as a predictive biomarker for response to ICIs [155][98]. However, further scientific evidence is needed to clarify the similarities between PD-L1 detection on CTCs and tissue expression of PD-L1. Moreover, considering their rarity in the bloodstream, the utility of CTCs in immunotherapy is still in the early stages [156][99]. Nonetheless, emerging studies are documenting the role of extravesicles (EVs) as potential biomarkers for immunotherapy. Therefore, EV-based liquid biopsies could eventually identify tumor-expressed proteins, DNA mutations, RNA landscape, and T-cell reactivity in patients under treatment with ICIs [157][100]. RAS assessment in mCRC is essential to select patients suitable for anti-EGFR therapy. The concordance between tissue detection and somatic mutations detected in ctDNA appeared high in patients with advanced tumors, supporting blood-based testing. Moreover, ctDNA was shown to be highly useful for monitoring treatment response. However, some clinicopathological features, including tumor histology (mucinous) and metastatic sites (peritoneal, lung), negatively influenced RAS detection in ctDNA [158][101]. Along with RAS mutation, TP53 mutations were widely detected in the ctDNA of CRC patients, with a high correlation between tissue and plasma detection. In CRC patients who did not progress to metastatic disease after primary surgery, the VAF for TP53 mutations decreased. By contrast, increased levels were associated with the development of liver metastasis [159][102]. Moreover, TP53 mutations were significantly correlated with increased VEGFA mRNA tissue expression, suggesting that these patients are expected to benefit from anti-VEGF therapy [160][103]. Nonetheless, TP53 mutations might occur as a consequence of several treatment strategies. In CRC, these mutations were particularly linked to cetuximab therapy, leading to resistant clones, and, therefore, influencing treatment opportunities [161][104]. Immunotherapy and targeted therapy are major therapeutic breakthroughs in cancer care, and one of the most challenging concerns is proper patient selection. To overcome these shortcomings, liquid biopsy-based biomarkers represent a promising tool, hence they require detection methods with sufficient specificity, sensitivity and predictive power [162][105].4. Remaining Obstacles in Clinical Applications of Liquid Biopsy
Despite many pieces of scientific evidence highlighting the potential benefits of liquid biopsies in cancer care, numerous limitations remain for their clinical use. CTCs have great potential as diagnostic, prognostic, predictive, as well as monitoring tools. However, their translation into clinical practice is still restricted amid their isolation from the bloodstream [163][106]. To correctly identify and analyze CTCs, it is essential to understand the obstacles surrounding their use. An important challenge is their extreme rarity, which makes them hard to locate. Among blood cells, CTCs are considered to be one in a million, or a billion [164][107]. Moreover, their concentration is much lower in the early stages compared to metastatic disease. Another important issue regards the size, physical characteristics and complexity of surface protein expression [165][108]. The main techniques used for CTCs enrichment are antigen-dependent (positive or negative selection), antigen-independent, or a combination of both [166][109]. In the case of positive selection, the efficacy of CTCs affinity is mainly influenced by antibody selectivity in the enrichment process. In this regard, the antibodies utilized may suffer from low selectivity as the target cell-surface proteins could also be expressed on other cells. Antibody cocktails targeting several cell-surface proteins have been used to overcome this limitation [166][109]. Moreover, the negative enrichment methods using magnetic bands to deplete circulating platelets and leucocytes have also been used to overcome the limitations of positive enrichment [167][110]. While using antigen-independent methods, the isolation of CTCs depends on the electric charge, density, size and deformability. However, with these techniques, CTCs purity is usually low due to size overlapping with WBC [168][111]. For exosomal segregation, ultracentrifugation is thought to be the most efficient method. Although convenient and requiring reasonable costs, ultracentrifugation encounters several limitations. A significant concern is the co-purification of lipoproteins and protein aggregates along with EVs [169][112]. Hence, it could be overcome by combining ultracentrifugation with density-gradient mechanisms. However, the structures with similar densities are indistinguishable. Immune isolation targets EVs with a particular surface marker representing a more specific method. Nonetheless, the targeted surface marker could also be found on other EV subsets [170][113]. ctDNA is a small fraction representing about 0.01% of cfDNA. One of the main logistical reasons limiting the extensive use of cDNA-based analysis is represented by their feasibility outside the academic cancer centers [171][114]. The available techniques for cDNA detection are based on PCR and NGS. These techniques were updated over time to better fit the low concentrations in the bloodstream. However, despite their sensitivity, PCR-based assays are limited by a low multiplexing capacity that permits the analysis of a small number of loci [172][115]. On the other hand, the sensitivity of NGS-based assays is low and inversely proportional to the examined loci [163, 173][106][116]. Another concerning issue implies the predictive value of a small set of mutations which could also be found in healthy individuals due to clonal hematopoiesis [173][116]. Moreover, the preanalytical sample preparation of cDNA lacks standardization limiting its implementation into clinical practice. Currently, a significant limitation preventing cDNA molecular panels in CRC is the lack of precise data showing that liquid biopsy findings could drive the therapeutic approach [174][117]. Similar to cDNA, the most relevant limitation in cRNA analysis is represented by preanalytical and analytical phases, along with a lack of standard extraction protocols [175][118]. A critical issue implies the hemolytic process, which occurs during extraction and preparation and can influence the levels of detected miRNAs. For this reason, monitoring the hemolysis of all samples in a pre-analytical phase is mandatory [176][119]. Another essential constraint regards residual platelets and microparticles resulting from plasma processing that can influence miRNA measurements [177][120]. Moreover, it is difficult to determine if the levels of plasma miRNA are confounded by comorbidities or are cancer-related. Therefore, a significant challenge is establishing which body fluid detection method is the most appropriate for CRC screening [78][121]. Even if several logistic and biological limitations are encountered at the present moment, liquid biopsies will more likely become a fundamental tool in the management of CRC patients in all stages of disease-related interventions. Evaluation of ctDNA levels in patients with stage II CRC has led to a decrease in adjuvant chemotherapy administration while maintaining a favorable recurrence-free survival (DYNAMIC II study) [178][122]. Additionally, the ongoing DYNAMIC III study attempts to evaluate a ctDNA-guided treatment approach in the setting of stage III CRC [179][123]. Liquid biopsy also plays an important role in assessing minimal residual disease, guiding the timing of therapeutic interventions based on ctDNA levels obtained at crucial points during treatment [180][124]. Furthermore, extensive research has proved that liquid biopsy offers strong indicators of early disease recurrence, with a significant median lead time of 8.7 months over conventional assessment methods [181][125]. Liquid biopsies have demonstrated valuable clinical application in detecting patients with acquired resistance to anti-EGFR therapy, therefore allowing a better selection of cases who may benefit from an EGFR rechallenge [182][126]. For CRC early diagnosis, however, comprehensive studies comparing the available data on liquid biopsy to current screening methods are necessary in order to find the most advantageous clinical use in everyday practice.
5. ConclusionsConclusions
Despite the implementation of extensive programs designed to diagnose CRC in the early stages, the adherence of the general population to screening protocols remains unsatisfactory. Liquid biopsy comes as a novel, minimally invasive tool, adequate for early diagnosis of malignancy while also attempting to overcome the limitation showed by traditional CRC screening and tissue sampling methods. Research has proved that liquid biopsy analytes and biomarkers offer valuable information regarding carcinogenesis and could indeed single out individuals at risk for developing CRC or those presenting early-stage CRC otherwise undetected during conventional screening tests. Since FDA has taken the necessary steps to approve CellSearch System for CTC detection and enhancement during clinical studies, the scientific community could soon implement the test in everyday clinical practice. In addition, studies have identified numerous methylation markers indicative of malignancy. In this regard, the detection of SEPT9 gene methylation in DNA fragments derivative from tumor masses is the most widely explored sequence and therefore experiences more promising results. As a result of thorough research, the SEPT9 methylation model comes within reach of validation for current practice, with tests such as Epi proColon expressing significant diagnostic values for CRC early detection. Several extensively researched miRNAs are also approaching clinical utility in everyday practice. miRNA-92a proves relevant sensitivity and specificity values in detecting early CRC cases, as well as advanced adenomas, while panels including multiple miRNAs show improved accuracy when compared to single markers. Additionally, exosomal miRNA-23a has been shown to differentiate patients with early-stage disease with high accuracy, indicating that protocols defining its clinical applicability could be imminent. However, in order to successfully isolate compelling biomarkers, blood samples require complex manipulation techniques that often show unsatisfactory efficiency and prove to be time-consuming and costly. Studies conducted thus far have demonstrated encouraging accuracy, yet further research is critical in order to validate liquid biopsy as a screening and early diagnostic technique. Furthermore, the utility of liquid biopsies in the era of precision medicine is an important frontier that demands thorough inquiry.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Hull, R.; Francies, F.Z.; Oyomno, M.; Dlamini, Z. Colorectal Cancer Genetics, Incidence and Risk Factors: In Search for Targeted Therapies. Cancer Manag. Res. 2020, 12, 9869–9882.
- Thanikachalam, K.; Khan, G. Colorectal Cancer and Nutrition. Nutrients 2019, 11, 164.
- Ollberding, N.J.; Nomura, A.M.; Wilkens, L.R.; Henderson, B.E.; Kolonel, L.N. Racial/ethnic differences in colorectal cancer risk: The multiethnic cohort study. Int. J. Cancer 2011, 129, 1899–1906.
- Simon, K. Colorectal cancer development and advances in screening. Clin. Interv. Aging 2016, 11, 967–976.
- Gunderson, L.L.; Jessup, J.M.; Sargent, D.J.; Greene, F.L.; Stewart, A.K. Revised TN categorization for colon cancer based on national survival outcomes data. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 264–271.
- Ladabaum, U.; Dominitz, J.A.; Kahi, C.; Schoen, R.E. Strategies for Colorectal Cancer Screening. Gastroenterology 2020, 158, 418–432.
- Issa, I.A.; Noureddine, M. Colorectal cancer screening: An updated review of the available options. World J. Gastroenterol. 2017, 23, 5086–5096.
- Mannucci, A.; Zuppardo, R.A.; Rosati, R.; Leo, M.D.; Perea, J.; Cavestro, G.M. Colorectal cancer screening from 45 years of age: Thesis, antithesis and synthesis. World J. Gastroenterol. 2019, 25, 2565–2580.
- Ahmed, M. Colon Cancer: A Clinician’s Perspective in 2019. Gastroenterol. Res. 2020, 13, 1–10.
- Binefa, G.; Rodríguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014, 20, 6786–6808.
- Ding, Y.; Li, W.; Wang, K.; Xu, C.; Hao, M.; Ding, L. Perspectives of the Application of Liquid Biopsy in Colorectal Cancer. BioMed Res. Int. 2020, 2020, 6843180.
- Burz, C.; Rosca, A.; Pop, V.V.; Buiga, R.; Aldea, C.; Samasca, G.; Silaghi, C.; Sur, D.; Lupan, I.; Pricopie, A. Liquid biopsy challenge and hope in colorectal cancer. Expert Rev. Mol. Diagn. 2019, 19, 341–348.
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548.
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841.
- He, J.; Xi, N.; Han, Z.; Luo, W.; Shen, J.; Wang, S.; Li, J.; Guo, Z.; Cheng, H. The Role of Liquid Biopsy Analytes in Diagnosis, Treatment and Prognosis of Colorectal Cancer. Front. Endocrinol. 2022, 13, 875442.
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415.
- de Wit, S.; van Dalum, G.; Terstappen, L.W. Detection of circulating tumor cells. Scientifica 2014, 2014, 819362.
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 404.
- Mohan, S.; Chemi, F.; Brady, G. Challenges and unanswered questions for the next decade of circulating tumour cell research in lung cancer. Transl. Lung Cancer Res. 2017, 6, 454–472.
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.I.; Morgan, B.; Trautwein, J.; et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra47.
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 8152–8162.
- Danese, E.; Montagnana, M.; Lippi, G. Circulating molecular biomarkers for screening or early diagnosis of colorectal cancer: Which is ready for prime time? Ann. Transl. Med. 2019, 7, 610.
- Hardingham, J.E.; Grover, P.; Winter, M.; Hewett, P.J.; Price, T.J.; Thierry, B. Detection and Clinical Significance of Circulating Tumor Cells in Colorectal Cancer—20 Years of Progress. Mol. Med. 2015, 21 (Suppl. S1), S25–S31.
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424.
- Nicolazzo, C.; Colangelo, L.; Corsi, A.; Carpino, G.; Gradilone, A.; Sonato, C.; Raimondi, C.; Gaudio, E.; Gazzaniga, P.; Gianni, W. Liquid Biopsy in Rare Cancers: Lessons from Hemangiopericytoma. Anal. Cell. Pathol. 2018, 2018, 9718585.
- Han, S.; Zong, S.; Shi, Q.; Li, H.; Liu, S.; Yang, W.; Li, W.; Hou, F. Is Ep-CAM Expression a Diagnostic and Prognostic Biomarker for Colorectal Cancer? A Systematic Meta-Analysis. EBioMedicine 2017, 20, 61–69.
- Fehm, T.; Sagalowsky, A.; Clifford, E.; Beitsch, P.; Saboorian, H.; Euhus, D.; Meng, S.; Morrison, L.; Tucker, T.; Lane, N.; et al. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2002, 8, 2073–2084.
- Paoletti, C.; Larios, J.M.; Muñiz, M.C.; Aung, K.; Cannell, E.M.; Darga, E.P.; Kidwell, K.M.; Thomas, D.G.; Tokudome, N.; Brown, M.E.; et al. Heterogeneous estrogen receptor expression in circulating tumor cells suggests diverse mechanisms of fulvestrant resistance. Mol. Oncol. 2016, 10, 1078–1085.
- Chu, C.H.; Liu, R.; Ozkaya-Ahmadov, T.; Swain, B.E.; Boya, M.; El-Rayes, B.; Akce, M.; Bilen, M.A.; Kucuk, O.; Sarioglu, A.F. Negative enrichment of circulating tumor cells from unmanipulated whole blood with a 3D printed device. Sci. Rep. 2021, 11, 20583.
- Maly, V.; Maly, O.; Kolostova, K.; Bobek, V. Circulating Tumor Cells in Diagnosis and Treatment of Lung Cancer. Vivo 2019, 33, 1027–1037.
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.; Schütze, K.; Capron, F.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am. J. Pathol. 2000, 156, 57–63.
- Bidard, F.C.; Proudhon, C.; Pierga, J.Y. Circulating tumor cells in breast cancer. Mol. Oncol. 2016, 10, 418–430.
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407.
- Tamminga, M.; Andree, K.C.; Hiltermann, T.; Jayat, M.; Schuuring, E.; van den Bos, H.; Spierings, D.; Lansdorp, P.M.; Timens, W.; Terstappen, L.; et al. Detection of Circulating Tumor Cells in the Diagnostic Leukapheresis Product of Non-Small-Cell Lung Cancer Patients Comparing CellSearch® and ISET. Cancers 2020, 12, 896.
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G.; et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250.
- Laget, S.; Broncy, L.; Hormigos, K.; Dhingra, D.M.; BenMohamed, F.; Capiod, T.; Osteras, M.; Farinelli, L.; Jackson, S.; Paterlini-Bréchot, P. Technical Insights into Highly Sensitive Isolation and Molecular Characterization of Fixed and Live Circulating Tumor Cells for Early Detection of Tumor Invasion. PLoS ONE 2017, 12, e0169427.
- Soler, A.; Cayrefourcq, L.; Mazel, M.; Alix-Panabières, C. EpCAM-Independent Enrichment and Detection of Viable Circulating Tumor Cells Using the EPISPOT Assay. Methods Mol. Biol. 2017, 1634, 263–276.
- Denève, E.; Riethdorf, S.; Ramos, J.; Nocca, D.; Coffy, A.; Daurès, J.P.; Maudelonde, T.; Fabre, J.M.; Pantel, K.; Alix-Panabières, C. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin. Chem. 2013, 59, 1384–1392.
- Abonnenc, M.; Manaresi, N.; Borgatti, M.; Medoro, G.; Fabbri, E.; Romani, A.; Altomare, L.; Tartagni, M.; Rizzo, R.; Baricordi, O. Programmable interactions of functionalized single bioparticles in a dielectrophoresis-based microarray chip. Anal. Chem. 2013, 85, 8219–8224.
- Sinkala, E.; Sollier-Christen, E.; Renier, C.; Rosàs-Canyelles, E.; Che, J.; Heirich, K.; Duncombe, T.A.; Vlassakis, J.; Yamauchi, K.A.; Huang, H.; et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 2017, 8, 14622.
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515.
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R.; et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016, 537, 102–106.
- Gasch, C.; Plummer, P.N.; Jovanovic, L.; McInnes, L.M.; Wescott, D.; Saunders, C.M.; Schneeweiss, A.; Wallwiener, M.; Nelson, C.; Spring, K.J.; et al. Heterogeneity of miR-10b expression in circulating tumor cells. Sci. Rep. 2015, 5, 15980.
- Babayan, A.; Alawi, M.; Gormley, M.; Müller, V.; Wikman, H.; McMullin, R.P.; Smirnov, D.A.; Li, W.; Geffken, M.; Pantel, K.; et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 2016, 8, 56066–56080.
- Vasseur, A.; Kiavue, N.; Bidard, F.C.; Pierga, J.Y.; Cabel, L. Clinical utility of circulating tumor cells: An update. Mol. Oncol. 2021, 15, 1647–1666.
- Jiang, M.; Jin, S.; Han, J.; Li, T.; Shi, J.; Zhong, Q.; Li, W.; Tang, W.; Huang, Q.; Zong, H. Detection and clinical significance of circulating tumor cells in colorectal cancer. Biomark. Res. 2021, 9, 85.
- Tsai, W.S.; Nimgaonkar, A.; Segurado, O.; Chang, Y.; Hsieh, B.; Shao, H.J.; Wu, J.C.; Lai, J.M.; Javey, M.; Watson, D.; et al. Prospective clinical study of circulating tumor cells for colorectal cancer screening. J. Clin. Oncol. 2018, 36 (Suppl. S4), 556.
- Cima, I.; Kong, S.L.; Sengupta, D.; Tan, I.B.; Phyo, W.M.; Lee, D.; Hu, M.; Iliescu, C.; Alexander, I.; Goh, W.L.; et al. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci. Transl. Med. 2016, 8, 345ra89.
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Xu, H.; Wang, Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer 2015, 15, 202.
- Bidard, F.C.; Kiavue, N.; Ychou, M.; Cabel, L.; Stern, M.H.; Madic, J.; Saliou, A.; Rampanou, A.; Decraene, C.; Bouché, O.; et al. Circulating Tumor Cells and Circulating Tumor DNA Detection in Potentially Resectable Metastatic Colorectal Cancer: A Prospective Ancillary Study to the Unicancer Prodige-14 Trial. Cells 2019, 8, 516.
- Aranda, E.; Viéitez, J.M.; Gómez-España, A.; Gil Calle, S.; Salud-Salvia, A.; Graña, B.; Garcia-Alfonso, P.; Rivera, F.; Quintero-Aldana, G.A.; Reina-Zoilo, J.J.; et al. FOLFOXIRI plus bevacizumab versus FOLFOX plus bevacizumab for patients with metastatic colorectal cancer and ≥3 circulating tumour cells: The randomised phase III VISNÚ-1 trial. ESMO Open 2020, 5, e000944.
- Rodriguez-Casanova, A.; Costa-Fraga, N.; Bao-Caamano, A.; López-López, R.; Muinelo-Romay, L.; Diaz-Lagares, A. Epigenetic Landscape of Liquid Biopsy in Colorectal Cancer. Front. Cell Dev. Biol. 2021, 9, 622459.
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1649–1683.
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045.
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24.
- Zhang, H.; Wang, L.; Li, C.; Yu, Y.; Yi, Y.; Wang, J.; Chen, D. Exosome-Induced Regulation in Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 1464.
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. 2021, 11, 2783–2797.
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547.
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta. Rev. Cancer 2019, 1871, 455–468.
- Munson, P.; Shukla, A. Exosomes: Potential in Cancer Diagnosis and Therapy. Medicines 2015, 2, 310–327.
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215.
- Sur, D.; Coza, O.; Havasi, A.; Cainap, C.; Burz, C.; Vlad, C.; Balacescu, O.; Alexandru, I.; Lisencu, C. Exosomal miRNAs in colorectal cancer: The carriers of useful news. J. BU ON. Off. J. Balk. Union Oncol. 2020, 25, 23–34.
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. CCS 2021, 19, 47.
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437.
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75.
- Yu, D.D.; Wu, Y.; Shen, H.Y.; Lv, M.M.; Chen, W.X.; Zhang, X.H.; Zhong, S.L.; Tang, J.H.; Zhao, J.H. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015, 106, 959–964.
- Hu, G.; Drescher, K.M.; Chen, X.M. Exosomal miRNAs: Biological Properties and Therapeutic Potential. Front. Genet. 2012, 3, 56.
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3-receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 2017, 7, 1333–1345.
- Saad, M.G.; Beyenal, H.; Dong, W.J. Exosomes as Powerful Engines in Cancer: Isolation, Characterization and Detection Techniques. Biosensors 2021, 11, 518.
- Jung, M.K.; Mun, J.Y. Sample Preparation and Imaging of Exosomes by Transmission Electron Microscopy. J. Vis. Exp. JoVE 2018, 131, e56482.
- McNicholas, K.; Michael, M.Z. Immuno-characterization of Exosomes Using Nanoparticle Tracking Analysis. Methods Mol. Biol. 2017, 1545, 35–42.
- Mihály, J.; Deák, R.; Szigyártó, I.C.; Bóta, A.; Beke-Somfai, T.; Varga, Z. Characterization of extracellular vesicles by IR spectroscopy: Fast and simple classification based on amide and CH stretching vibrations. Biochim. Biophys. Acta. Biomembr. 2017, 1859, 459–466.
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921.
- Karimi, N.; Ali Hosseinpour Feizi, M.; Safaralizadeh, R.; Hashemzadeh, S.; Baradaran, B.; Shokouhi, B.; Teimourian, S. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J. Chin. Med. Assoc. JCMA 2019, 82, 215–220.
- Yan, S.; Jiang, Y.; Liang, C.; Cheng, M.; Jin, C.; Duan, Q.; Xu, D.; Yang, L.; Zhang, X.; Ren, B.; et al. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J. Cell. Biochem. 2018, 119, 4113–4119.
- Liu, X.; Chen, X.; Zeng, K.; Xu, M.; He, B.; Pan, Y.; Sun, H.; Pan, B.; Xu, X.; Xu, T.; et al. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018, 9, 1037.
- Wang, J.; Yan, F.; Zhao, Q.; Zhan, F.; Wang, R.; Wang, L.; Zhang, Y.; Huang, X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 2017, 7, 4150.
- Zou, S.L.; Chen, Y.L.; Ge, Z.Z.; Qu, Y.Y.; Cao, Y.; Kang, Z.X. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark. 2019, 26, 69–77.
- Min, L.; Chen, L.; Liu, S.; Yu, Y.; Guo, Q.; Li, P.; Zhu, S. Loss of Circulating Exosomal miR-92b is a Novel Biomarker of Colorectal Cancer at Early Stage. Int. J. Med. Sci. 2019, 16, 1231–1237.
- Ren, D.; Lin, B.; Zhang, X.; Peng, Y.; Ye, Z.; Ma, Y.; Liang, Y.; Cao, L.; Li, X.; Li, R.; et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 2017, 8, 49807–49823.
- Liu, W.; Yang, D.; Chen, L.; Liu, Q.; Wang, W.; Yang, Z.; Shang, A.; Quan, W.; Li, D. Plasma Exosomal miRNA-139-3p is a Novel Biomarker of Colorectal Cancer. J. Cancer 2020, 11, 4899–4906.
- Liu, X.; Pan, B.; Sun, L.; Chen, X.; Zeng, K.; Hu, X.; Xu, T.; Xu, M.; Wang, S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 746–754.
- Cui, X.; Lv, Z.; Ding, H.; Xing, C.; Yuan, Y. MiR-1539 and Its Potential Role as a Novel Biomarker for Colorectal Cancer. Front. Oncol. 2021, 10, 531244.
- Zhu, M.; Huang, Z.; Zhu, D.; Zhou, X.; Shan, X.; Qi, L.W.; Wu, L.; Cheng, W.; Zhu, J.; Zhang, L.; et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017, 8, 17081–17091.
- Cheng, W.C.; Liao, T.T.; Lin, C.C.; Yuan, L.E.; Lan, H.Y.; Lin, H.H.; Teng, H.W.; Chang, H.C.; Lin, C.H.; Yang, C.Y.; et al. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int. J. Cancer 2019, 145, 2209–2224.
- Yan, S.; Han, B.; Gao, S.; Wang, X.; Wang, Z.; Wang, F.; Zhang, J.; Xu, D.; Sun, B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017, 8, 60149–60158.
- Buchler, T. Microsatellite Instability and Metastatic Colorectal Cancer—A Clinical Perspective. Front. Oncol. 2022, 12, 888181.
- Gorzo, A.; Galos, D.; Volovat, S.R.; Lungulescu, C.V.; Burz, C.; Sur, D. Landscape of Immunotherapy Options for Colorectal Cancer: Current Knowledge and Future Perspectives beyond Immune Checkpoint Blockade. Life 2022, 12, 229.
- Carlsen, L.; Huntington, K.E.; El-Deiry, W.S. Immunotherapy for Colorectal Cancer: Mechanisms and Predictive Biomarkers. Cancers 2022, 14, 1028.
- Thompson, J.R.; Menon, S.P. Liquid Biopsies and Cancer Immunotherapy. Cancer J. 2018, 24, 78–83.
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; van der Heijden, M.S.; et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1842–1853.
- Bratman, S.V.; Yang, S.; Iafolla, M.; Liu, Z.; Hansen, A.R.; Bedard, P.L.; Lheureux, S.; Spreafico, A.; Razak, A.A.; Shchegrova, S.; et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer 2020, 1, 873–881.
- Jiao, X.; Wei, X.; Li, S.; Liu, C.; Chen, H.; Gong, J.; Li, J.; Zhang, X.; Wang, X.; Peng, Z.; et al. A genomic mutation signature predicts the clinical outcomes of immunotherapy and characterizes immunophenotypes in gastrointestinal cancer. NPJ Precis. Oncol. 2021, 5, 36.
- Passaro, A.; Stenzinger, A.; Peters, S. Tumor Mutational Burden as a Pan-cancer Biomarker for Immunotherapy: The Limits and Potential for Convergence. Cancer Cell 2020, 38, 624–625.
- Wu, X.; Zhu, L.; Ma, P.C. Next-Generation Novel Noninvasive Cancer Molecular Diagnostics Platforms Beyond Tissues. Am. Soc. Clin. Oncol. Educ. Book. 2018, 38, 964–977.
- Friedlaender, A.; Nouspikel, T.; Christinat, Y.; Ho, L.; McKee, T.; Addeo, A. Tissue-Plasma TMB Comparison and Plasma TMB Monitoring in Patients With Metastatic Non-small Cell Lung Cancer Receiving Immune Checkpoint Inhibitors. Front. Oncol. 2020, 10, 142.
- Dall’Olio, F.G.; Gelsomino, F.; Conci, N.; Marcolin, L.; De Giglio, A.; Grilli, G.; Sperandi, F.; Fontana, F.; Terracciano, M.; Fragomeno, B.; et al. PD-L1 Expression in Circulating Tumor Cells as a Promising Prognostic Biomarker in Advanced Non-small-cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Clin. Lung Cancer 2021, 22, 423–431.
- Fatima, S.; Ma, Y.; Safrachi, A.; Haider, S.; Spring, K.J.; Vafaee, F.; Scott, K.F.; Roberts, T.L.; Becker, T.M.; de Souza, P. Harnessing Liquid Biopsies to Guide Immune Checkpoint Inhibitor Therapy. Cancers 2022, 14, 1669.
- Mathew, M.; Zade, M.; Mezghani, N.; Patel, R.; Wang, Y.; Momen-Heravi, F. Extracellular Vesicles as Biomarkers in Cancer Immunotherapy. Cancers 2020, 12, 2825.
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodríguez, C.; Brozos, E.; et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1325–1332.
- Khan, Z.A.; Jonas, S.K.; Le-Marer, N.; Patel, H.; Wharton, R.Q.; Tarragona, A.; Ivison, A.; Allen-Mersh, T.G. P53 mutations in primary and metastatic tumors and circulating tumor cells from colorectal carcinoma patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 3499–3504.
- Li, A.M.; Boichard, A.; Kurzrock, R. Mutated TP53 is a marker of increased VEGF expression: Analysis of 7525 pan-cancer tissues. Cancer Biol. Ther. 2020, 21, 95–100.
- Ghatalia, P.; Smith, C.H.; Winer, A.; Gou, J.; Kiedrowski, L.A.; Slifker, M.; Saltzberg, P.D.; Bubes, N.; Anari, F.M.; Kasireddy, V.; et al. Clinical Utilization Pattern of Liquid Biopsies (LB) to Detect Actionable Driver Mutations, Guide Treatment Decisions and Monitor Disease Burden During Treatment of 33 Metastatic Colorectal Cancer (mCRC) Patients (pts) at a Fox Chase Cancer Center GI Oncology Subspecialty Clinic. Front. Oncol. 2019, 8, 652.
- Taefehshokr, S.; Parhizkar, A.; Hayati, S.; Mousapour, M.; Mahmoudpour, A.; Eleid, L.; Rahmanpour, D.; Fattahi, S.; Shabani, H.; Taefehshokr, N. Cancer immunotherapy: Challenges and limitations. Pathol. Res. Pract. 2022, 229, 153723.
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186.
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078.
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877.
- Shen, Z.; Wu, A.; Chen, X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 2017, 46, 2038–2056.
- Liu, Z.; Fusi, A.; Klopocki, E.; Schmittel, A.; Tinhofer, I.; Nonnenmacher, A.; Keilholz, U. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J. Transl. Med. 2011, 9, 70.
- Habli, Z.; AlChamaa, W.; Saab, R.; Kadara, H.; Khraiche, M.L. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers 2020, 12, 1930.
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10.
- Wang, W.; Luo, J.; Wang, S. Recent Progress in Isolation and Detection of Extracellular Vesicles for Cancer Diagnostics. Adv. Healthc. Mater. 2018, 7, e1800484.
- Downing, A.; Morris, E.J.; Corrigan, N.; Sebag-Montefiore, D.; Finan, P.J.; Thomas, J.D.; Chapman, M.; Hamilton, R.; Campbell, H.; Cameron, D.; et al. High hospital research participation and improved colorectal cancer survival outcomes: A population-based study. Gut 2017, 66, 89–96.
- Wan, J.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238.
- Domínguez-Vigil, I.G.; Moreno-Martínez, A.K.; Wang, J.Y.; Roehrl, M.; Barrera-Saldaña, H.A. The dawn of the liquid biopsy in the fight against cancer. Oncotarget 2017, 9, 2912–2922.
- Mauri, G.; Vitiello, P.P.; Sogari, A.; Crisafulli, G.; Sartore-Bianchi, A.; Marsoni, S.; Siena, S.; Bardelli, A. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br. J. Cancer 2022, 127, 394–407.
- Sourvinou, I.S.; Markou, A.; Lianidou, E.S. Quantification of circulating miRNAs in plasma: Effect of preanalytical and analytical parameters on their isolation and stability. J. Mol. Diagn. 2013, 15, 827–834.
- Yang, J.; Ma, D.; Fesler, A.; Zhai, H.; Leamniramit, A.; Li, W.; Wu, S.; Ju, J. Expression analysis of microRNA as prognostic biomarkers in colorectal cancer. Oncotarget 2016, 8, 52403–52412.
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE 2013, 8, e64795.
- Marcuello, M.; Vymetalkova, V.; Neves, R.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flügen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019, 69, 107–122.
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272.
- Malla, M.; Loree, J.M.; Kasi, P.M.; Parikh, A.R. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2022, 40, 2846–2857.
- Heidrich, I.; Abdalla, T.; Reeh, M.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as a Liquid Biopsy Marker in Colorectal Cancer. Cancers 2021, 13, 4500.
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131.
- Cremolini, C.; Rossini, D.; Dell’Aquila, E.; Lonardi, S.; Conca, E.; Del Re, M.; Busico, A.; Pietrantonio, F.; Danesi, R.; Aprile, G.; et al. Rechallenge for Patients With RAS and BRAF Wild-Type Metastatic Colorectal Cancer With Acquired Resistance to First-line Cetuximab and Irinotecan: A Phase 2 Single-Arm Clinical Trial. JAMA Oncol. 2019, 5, 343–350.
