Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Robin A J Nicholas.
Mycoplasma bovis is a cause of bronchopneumonia, mastitis and arthritis but may also affect other main organs in cattle such us the eye, ear or brain. M. bovis infections are responsible for substantial economic health and welfare problems worldwide. M. bovis has spread worldwide, including to countries for a long time considered free of the pathogen.
- Mycoplasma bovis
- cattle
- disease
- prevalence
- control
1. Introduction
In 2017, New Zealand became the last of the major cattle-rearing countries to be infected with Mycoplasma bovis [1]. Finland had also remained free until relatively recently but became infected via imported cattle in 2012 [2]. Undoubtedly, M. bovis is now the most important mycoplasma of livestock being a primary cause of mastitis, arthritis, keratoconjunctivitis and other disorders as well as a major player in the bovine respiratory disease complex (BRD) [3]. Previously Mycoplasma mycoides subsp. mycoides, the aetiological agent of the World Organisation for Animal Health (OIE)-listed contagious bovine pleuropneumonia, had this dubious distinction but this mycoplasma is now confined to countries in sub Saharan Africa.
Mycoplasma bovis was first reported in the USA in 1961 from a case of bovine mastitis then was probably exported in cattle of high genetic quality to Israel [3]. It then spread around the world, reaching the UK and the rest of Europe in the mid1970s (Figure 1). International trade in cattle and cattle products like semen has enabled its silent spread to all continents where cattle are kept. The date of isolation in a particular country, of course, is not necessarily the date of introduction even in the USA as mycoplasmas were very much an unknown quantity and their fastidious nature made isolation and detection an extremely difficult task. Indeed, it has only been in the last two decades with the introduction of DNA amplification techniques that detection and identification have become routine in many parts of the world. However, not all countries have veterinary diagnostic laboratories which can identify these organisms.
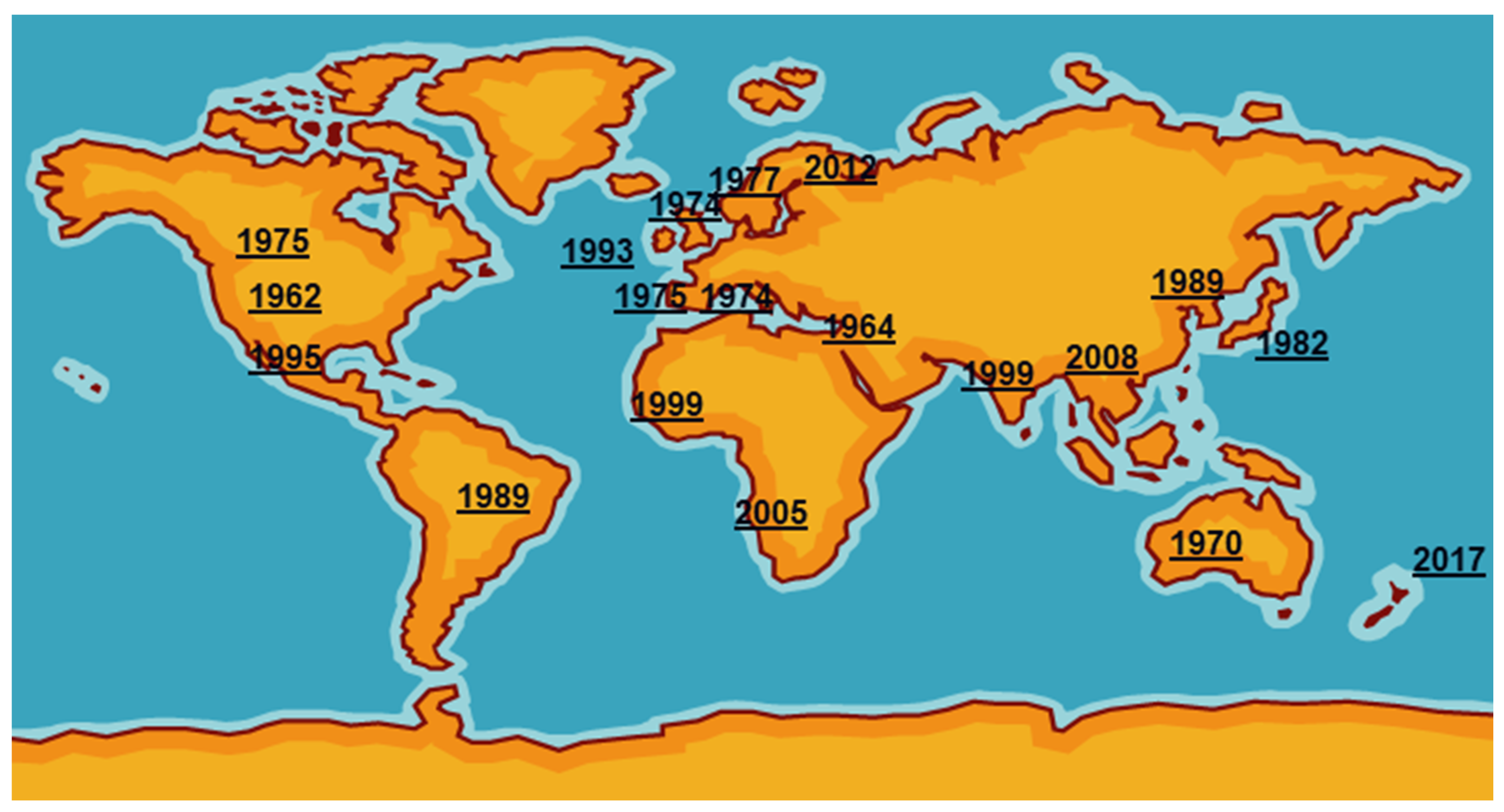
Figure 1.
First detections of
Mycoplasma bovis
around the world.
Initially the importance of M. bovis, particularly in BRD, was underestimated because of the promotion of more established and easier detectable organisms like the bacteria Mannheimia haemolytica, Histophilus somni and Pasteurella multocida and viruses, namely bovine respiratory syncytial disease, parainfluenza-3 virus, bovine herpesviruses, coronaviruses and bovine viral diarrhoea virus. The presence of M. bovis in healthy cattle, although at a much lower levels than infected ones, delayed recognition of its pathogenicity. Once the importance of environmental factors such as weather, variation in strain virulence and its interaction with the BRD pathogens were known, studies quickly demonstrated its widespread prevalence in pneumonic calves and, later, older cattle.
Despite attempts going back nearly half a century, control of M. bovis diseases is still problematic because of a lack of an effective commercial vaccine. Many have been marketed, particularly in the USA, but little data exist to assess their immunogenicity and protective properties [4]. To be valuable they are required to be part of multivalent vaccines incorporating the causative bacteria and viruses currently available for BRD. Presently, no vaccine is available for mycoplasma mastitis, a major problem in large dairy herds of North America where they are often untreatable. Indeed, the major trend in the last two decades has been the alarming decrease in susceptibility of M. bovis to the commonly used antimicrobials including the fluoroquinolones [5].
2. Currently Used Diagnostic Methods
The clinical signs of infections in cattle associated with M. bovis are non-specific; for that reason, sensitive, accurate and rapid testing of animals is needed for reliable diagnosis. Culturing of M. bovis is a gold standard method but is time-consuming and requires specific conditions. Different kinds of media are widely used in experimental studies and in confirmation of infection caused by M. bovis, and include Hayflick’s [42][6], modified PPLO [43][7] and Eaton’s [44][8]. Mycoplasmas are fastidious, slow growing and can be easily overgrown by other bacteria. During the last few years various tests have been used for the detection of M. bovis infections in cattle.2.1. Real-Time PCR Assays for M. bovis Detection
2.2. Fast and Cost-Effective Assays for M. bovis Detection
Another approach for M. bovis detection is to design a simple and cost-effective assay run at a single temperature without the need of using specific equipment, which will be useful to process in developing countries. LAMP (loop-mediated Isothermal amplification) is recently of interest because it enables results to be received quickly, and the reaction is normally completed in less than 2 h; furthermore, there is no need to have expensive laboratory equipment, as it is performed at a single temperature [57][18]. LAMP gives better results than qPCR when performed on purified DNA but is susceptible to contamination. Two assays, namely LAMP and qPCR developed for M. bovis detection in milk samples from individual cow quarters and bulk tank milk samples, accurately detected M. bovis isolates but gave false positive results for one Mycoplasma bovigenitalium isolate [47][16]. Another method called isothermal DNA amplification assay, a technique based on recombinase polymerase amplification (RPA) with lateral flow dipstick (LFD), allows one to obtain the result in 30 min and is dedicated for M. bovis DNA extracted directly from clinical samples i.e., nasal swabs, lungs tissue samples, joint fluids and bulk tank milk samples; no cross-reactions were observed with other Mycoplasma species [53][19]. Usually, LAMP assays are more sensitive than end-point PCRs, for example high sensitivity and specificity for all milk sample types was obtained with the use of LAMP combined with a procedure for ultra-rapid extraction (PURE-LAMP), in which various sample types i.e., bulk tank milk, mature milk, colostrum/transitional milk and mastitis milk were examined [52][20]. Similar parameters were obtained in LAMP for the examination of M. bovis in milk from mastitis cases [51][21].2.3. Immunohistochemistry and In-Situ Hybridization
Although molecular methods are advantageous, they can only provide the data on M. bovis DNA, and there is lacking information about the presence of viable bacteria and their associaation with the lesion. Immunohistochemistry (IHC) and in-situ hybridization (ISH) are types of techniques which have the advantage that they are able to detect the localization of M. bovis antigen or DNA, respectively, in the examined tissue of the infected animals [12,19,41,58,59][22][23][24][25][26]. The IHC used in the study on calves experimentally infected with M. bovis allows one to detect M. bovis antigen in the bronchiolar epithelial cells in the lung tissue with histopathological changes that are characteristic for bronchiolitis [19][23]. Results of another experiment proved that M. bovis antigen was detected on the surface and inside the cytoplasm of bronchiolar epithelial cells in the pneumonic foci and in the cytoplasm of phagocytes at the margin of bronchiolar exudates [58][25]. In the study on aborted foetus and neonatal calf that were infected with M. bovis, its antigen was found with the use of IHC in the brain, liver, lungs and placenta of aborted foetus, and ISH showed the presence of its DNA i.e., in lungs and placenta of the examined animals [41][24]. The research on long-term survival of M. bovis in tissues of infected calves showed the persistence of this pathogen in necrotic lung lesions several weeks after the infection with the use of both methods [59][26]. It is also possible to examine the pulmonary samples of calves with BRD. IHC was used to detect the M. bovis antigen intralesional in different areas of the lungs [12][22]. However, while these techniques allow one to obtain significant information, they are also expensive, labour intensive and require trained staff.2.4. A Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for M. bovis Detection
The matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) procedure has been applied to M. bovis detection. It was optimised for the detection of M. bovis isolates and found to be a suitable test for routine diagnostics in cattle, especially those from BRD cases. The protocol enables the identification of M. bovis from bronchoalveolar lavage fluid (BALF) after enrichment in culture. The higher number of positive samples was obtained after 72 h of enrichment. The main advantage of MALDI-TOF MS is that it only detects viable bacteria, which indicates that cattle have active rather than historic infections [54][27].2.5. Molecular Typing
The analysis of M. bovis isolates with typing and sequencing methods can give additional information about their relationships and evolution. The multilocus sequence typing (MLST) analysis was proved to be suitable for molecular typing of M. bovis and the assessment of geographical relatedness of isolates. The MLST scheme based on eleven housekeeping genes was evaluated. Three genes, dnaN, metS and hsp70, were taken for the sequence analysis and the remaining eight genes, i.e., adk, efp, gmk, gyrB, polC, rpoB, tpiA and uvrC were not chosen for the further analysis. It allows the acquiring of information on sequence variation, its type of distribution and disappearance of some sequence types [60][28]. A later study [61][29] assessed two MLST schemes for M. bovis isolate typing. The comparison of the performance of the two MLST schemes and additional identification of a new reference scheme capable of full typing of the examined isolates was made. The PubMLST reference method contains adh-1, gltX, gspA, gyrA, gyrB, pta-2, tdk and tkt locus; it is thought to be discriminatory and informative enough, but in this study, adh-1, one of the typing loci of M. bovis isolates, was missed. According to this reference scheme, the adh-1 locus should be retired from the analysis. This approach was not beneficial for the study because the discrimination index received with the use of the six remaining PubMLST loci failed to reach the benchmark recommended for a reference method, and the addition of a seventh locus had to be made. The alternative scheme contains seven loci: aptA, dnaA, metS, recA, rpoD, tkt and tufA. The comparisons of examined M. bovis genome sequences identified the dnaA locus from the alternative scheme as the optimal replacement for adh-1. Another approach for epidemiological studies is the use of whole genome sequencing (WGS) to evaluate the molecular epidemiology and genomic diversity of M. bovis isolates as well as their genetic relationship. The single nucleotide polymorphism (SNP) analysis can be used to assess the intraspecies relationship and the presence of a dominant genotype that can be associated with one type of disease. This study is relevant to better understand the global epidemiology of this important pathogen and to assess control strategies [62][30]. Comparison of the M. bovis sequences can be used in assessing the genetic diversity of the strains [63][31] or to get the information about gene virulence [64][32]. WGS was used in New Zealand to track the outbreaks first identified in 2017. In all, 171 isolates from 30 infected herds have so far been sequenced, and results indicate that the current outbreak was probably caused by recent entry of the mycoplasma, perhaps 1–2 years before detection, from a single source either as a single border crossing of a single clone or, potentially, up to three border crossings of three very closely related clones from the same source (TAG 2019) probably in germplasm imported from Europe.2.6. Serological Approaches
Serological diagnosis based on detection of specific antibodies to M. bovis is suitable and practical for the assessment of prevalence and epidemiological studies of herds [39][33]. Although serological testing is a reliable method for identification of infected animals, specific antibodies do not appear until 10 to 14 days after the infection but remain elevated for several months [65][34]. Various indirect ELISAs are used for anti-M. bovis antibody detection in cattle herds. The BIO K302 ELISA (BioX Diagnostics) was applied for evaluation of antibody response to M. bovis in serum and milk samples [13,66,67][35][36][37]. A study conducted in Belgium [67][37] showed that the ELISA is able to detect M. bovis specific antibodies in bulk tank milk up to 12 months after the outbreak of the disease. Researchers [66][36] examined bulk milk tank samples for all Danish herds with this ELISA and concluded that the cut-off value should be increased from 37%, as suggested for animal-level diagnosis, to 50%, to obtain more adequate sensitivity and specificity for bulk tank milk analysis. On the other hand, as a result of a European inter-laboratory comparison conducted on 180 serum samples, the sensitivity and specificity of BIO K302 ELISA was determined to be 49.1% and 89.6%, respectively [68][38]. However, in 2020 it was confirmed that this ELISA was suitable for the serological evaluation of anti-M. bovis antibodies in longitudinal studies. Despite the low number of apparent clinical mastitis cases, it was useful in evaluation of M. bovis seroprevalence in dairy herds, which was on average 38% (16–76%), as mentioned before [13][35]. Another indirect ELISA, made in-house and based on a fragment of a recombinant mycoplasma immunogenic lipase A (MilA), was developed [69][39]. This assay can be also useful for bulk tank milk sample analysis. The results of the presence of anti-M. bovis antibodies in bulk tank milk were positively correlated with the antibody detection in sera of the examined animals. Additionally, there was made a comparison between BIO K 260 (BioX Diagnostics) and the MilA ELISA [23][40], and the latter test gave a higher number of positive samples for M. bovis, and they were more convergent with those obtained with culture or real-time PCR. The obtained sensitivity and specificity for this test was 94.3% and 94.4%, respectively. Additionally, it was shown that the MilA ELISA is also suitable for testing the presence of anti-M. bovis antibodies in the early stages of calf life (from the 3rd week of life) [70][41].2.7. Interlaboratory Trials of Diagnostic Tests
M. bovis causes serious health problems in cattle herds almost all over the world, but its detection is not harmonised as yet and relies on different diagnostic methods, often in-house molecular techniques based on a variety of target genes and various different DNA extraction methods. There was conducted a European interlaboratory comparison of the diagnostic utility of the molecular tests for M. bovis detection [71][42]. Six laboratories from different countries were included in the study. Five different DNA extraction methods from bacterial culture and BALF samples were used. The molecular tests were made with the use of seven different PCR assays based on polC, oppD, uvrC and V4-V4 16S rRNA target genes. The comparison revealed that although the research used various assays, they had comparable diagnostic utility for M. bovis detection in cattle. The analytical specificity of the different PCR methods was comparable for all of the laboratories, except one, where M. agalactiae was detected because of the use of 16S rRNA target gene. The LOD was from 10 to 103 for the real-time, and from 103 to 106 CFU/mL for the end-point assays. According to the authors, this difference was acceptable. Cultures correctly detected the presence of M. bovis in bronchoalveolar lavage fluid samples and were consistent with PCR results. The recent comparison of diagnostic methods used in the different veterinary laboratories fortunately showed consensus.2.8. Mixed Infections
Other Mycoplasma spp. can also be associated with M. bovis infections in cattle. In BRD cases, most often M. dispar, M. canis and M. arginini are implicated [3,72][3][43]. In mastitis and reproductive disorders, M. bovigenitalium, M. californicum and M. alkalescens can also participate [73,74][44][45]. A test based on PCR with the 16SrRNA target gene and separation of the PCR products using denaturing gradient gel electrophoresis (PCR–DGGE) enabled the differentiation of 13 Mycoplasma spp. of bovine origin in mixed infections [75][46]. Traditionally, culture is used for the confirmation of BRD infections, but the incubation period for each examined bacterial pathogen is different and samples inoculated onto agar plates are often overgrown with other fast-growing bacteria. For that reason, the multiplex real-time PCRs used by some laboratories [49,50,76][15][47][48] are the most suitable for simultaneous direct detection of M. bovis and other pathogens involved in BRD, such as P. multocida, M. haemolytica and H. somni, in contrast to methods not dedicated for different pathogen identification in mixed infections such as one-target PCR, traditional culture or MALDI-TOF MS [77][49]. When using one target PCR, there is no information about the involvement of other pathogens in the disease, different bacteria have various growth requirements and slow growing bacteria can be easily overgrown by others, and MALDI-TOF MS is not able properly detect all organisms from polymicrobial samples. Various diagnostics methods for fast and accurate detection of M. bovis in various sample types and typing methods for identification and analysis of its strains in the last few years have been developed for evaluation of the disease course. Methods should be chosen according to the purpose of the survey, for herd-level testing or for individuals, or should be considered in terms of its usage for the specimen. The use of a combination of molecular, serological and culture-based methods is necessary for reliable diagnosis of diseases caused by this pathogen in cattle.References
- Ministry for Primary Industries. First Case of Mycoplasma Bovis Found in Bay of Plenty. 2017. Available online: https://www.tvnz.co.nz/one-news/new-zealand/first-case-mycoplasma-bovis-found-in-bay-plenty (accessed on 20 June 2020).
- Haapala, V.; Pohjanvirta, T.; Vähänikkilä, N.; Halkilahti, J.; Simonen, H.; Pelkonen, S.; Soveri, T.; Simojoki, H.; Autio, T. Semen as a source of Mycoplasma bovis mastitis in dairy herds. Vet. Microbiol. 2018, 216, 60–66.
- Nicholas, R.; Ayling, R.; McAuliffe, L. Mycoplasma Diseases of Ruminants, 1st ed.; CABI Publishing: Oxford, UK, 2008.
- Nicholas, R.A.J.; Fox, L.K.; Lysnyansky, I. Mycoplasma mastitis in cattle: To cull or not to cull. Vet. J. 2016, 216, 142–147.
- Klein, U.; de Jong, A.; Youala, M.; El Garch, F.; Stevenin, C.; Moyaert, H.; Rose, M.; Catania, S.; Gyuranecz, M.; Pridmore, A.; et al. New antimicrobial susceptibility data from monitoring of Mycoplasma bovis isolated in Europe. Vet. Microbiol. 2019, 238, 108432.
- Pfützner, H.; Sachse, K. Mycoplasma bovis as an agent of mastitis, pneumonia, arthritis and genital disorders in cattle. Rev. Sci. Tech. Off. Int. Epiz. 1996, 15, 1477–1494.
- Poumarat, F.; Longchambon, D.; Martel, J.L. Application of dot immunobinding on membrane filtration (MF dot) to the study of relationships within “M. mycoides cluster” and within “glucose and arginine-negative cluster” of ruminant mycoplasmas. Vet. Microbiol. 1992, 32, 375–390.
- Nicholas, R.A.J.; Baker, S.E. Recovery of mycoplasmas from animals. In Mycoplasma Protocols; Miles, R.J., Nicholas, R.A.J., Eds.; Humana Press: Totowa, NJ, USA, 1998; pp. 37–44.
- Thurmond, M.C.; Tyler, J.W.; Luiz, D.M.; Holmberg, C.A.; Picanso, J.P. The effect of pre-enrichment on recovery of Streptococcus agalactiae, Staphylococcus aureus and mycoplasma from bovine milk. Epidemiol. Infect. 1989, 103, 465–474.
- Behera, S.; Rana, R.; Gupta, P.K.; Kumar, D.; Sonal; Rekha, V.; Arun, T.R.; Jena, D. Development of real-time PCR assay for the detection of Mycoplasma bovis. Trop. Anim. Health Prod. 2018, 50, 875–882.
- Andrés-Lasheras, S.; Zaheer, R.; Ha, R.; Lee, C.; Jelinski, M.; McAllister, T.A. A direct qPCR screening approach to improve the efficiency of Mycoplasma bovis isolation in the frame of a broad surveillance study. J. Microbiol. Methods 2020, 169, 105805.
- Timonen, A.A.E.; Autio, T.; Pohjanvirta, T.; Häkkinen, L.; Katholm, J.; Petersen, A.; Mõtus, K.; Kalmus, P. Dynamics of the within-herd prevalence of Mycoplasma bovis intramammary infection in endemically infected dairy herds. Vet. Microbiol. 2020, 242, 108608.
- Gille, L.; Evrard, J.; Callens, J.; Supré, K.; Grégoire, F.; Boyen, F.; Haesebrouck, F.; Deprez, P.; Pardon, B. The presence of Mycoplasma bovis in colostrum. Vet. Res. 2020, 51, 54.
- Parker, A.M.; House, J.K.; Hazelton, M.S.; Bosward, K.L.; Sheehy, P.A. Comparison of culture and a multiplex probe PCR for identifying Mycoplasma species in bovine milk, semen and swab samples. PLoS ONE 2017, 12, e0173422.
- Pansri, P.; Katholm, J.; Krogh, K.M.; Aagaard, A.K.; Schmidt, L.M.B.; Kudirkiene, E.; Larsen, L.E.; Olsen, J.E. Evaluation of novel multiplex qPCR assays for diagnosis of pathogens associated with the bovine respiratory disease complex. Vet. J. 2020, 256, 105425.
- Appelt, S.; Aly, S.S.; Tonooka, K.; Glenn, K.; Xue, Z.; Lehenbauer, T.W.; Marco, M.L. Development and comparison of loop-mediated isothermal amplification and quantitative polymerase chain reaction assays for the detection of Mycoplasma bovis in milk. J. Dairy Sci. 2019, 102, 1985–1996.
- Punyapornwithaya, V.; Fox, L.K.; Gay, G.M.; Hancock, D.D.; Alldredge, J.R. Short communication: The effect of centrifugation and resuspension on the recovery of Mycoplasma species from milk. J. Dairy Sci. 2009, 92, 4444–4447.
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 2017, 107, 54–61.
- Zhao, G.; Hou, P.; Huan, Y.; He, C.; Wang, H.; He, H. Development of a recombinase polymerase amplification combined with a lateral flow dipstick assay for rapid detection of the Mycoplasma bovis. BMC Vet. Res. 2018, 14, 412.
- Itoh, M.; Hirano, Y.; Yamakawa, K.; Yasutomi, I.; Kuramoto, K.; Furuok, M.; Yamada, K. Combination of procedure for ultra rapid extraction (PURE) and loop-mediated isothermal amplification (LAMP) for rapid detection of Mycoplasma bovis in milk. J. Vet. Med. Sci. 2020, 19–0695.
- Ashraf, A.; Imran, M.; Yaqub, T.; Tayyab, M.; Shehzad, W.; Mingala, C.N.; Chang, Y.-F. Development and validation of a loop-mediated isothermal amplification assay for the detection of Mycoplasma bovis in mastitic milk. Folia Microb. 2018, 63, 373–380.
- Oliveira, T.E.S.; Pelaquim, I.F.; Flores, E.F.; Massi, R.P.; Valdiviezo, M.J.J.; Pretto-Giordano, L.G.; Alfieri, A.A.; Saut, J.P.E.; Headley, S.A. Mycoplasma bovis and viral agents associated with the development of bovine respiratory disease in adult dairy cows. Transbound. Emerg. Dis. 2019, 67, 82–93.
- Dudek, K.; Bednarek, D.; Ayling, R.D.; Kycko, A.; Reichert, M. Preliminary study on the effects of enrofloxacin, flunixin meglumine and pegbovigrastim on Mycoplasma bovis pneumonia. BMC Vet. Res. 2019, 15, 371.
- Hermeyer, K.; Peters, M.; Brügmann, M.; Jacobsen, B.; Hewicker-Trautwein, M. Demonstration of Mycoplasma bovis by immunohistochemistry and in situ hybridization in an aborted bovine fetus and neonatal calf. J. Vet. Diagn. Invest. 2012, 24, 364–369.
- Nunoya, T.; Omori, T.; Tomioka, H.; Umeda, F.; Suzuki, T.; Uetsuka, K. Intracellular Localization of Mycoplasma bovis in the Bronchiolar Epithelium of Experimentally Infected Calves. J. Comp. Path. 2020, 176, 14–18.
- Kleinschmidt, S.; Spergser, J.; Rosengarten, R.; Hewicker-Trautwein, M. Long-term survival of Mycoplasma bovis in necrotic lesions and in phagocytic cells as demonstrated by transmission and immunogold electron microscopy in lung tissue from experimentally infected calves. Vet. Microbiol. 2013, 162, 949–953.
- Bokma, J.; Van Driessche, L.; Deprez, P.; Haesebrouck, F.; Vahl, M.; Weesendorp, E.; Deurenberg, R.H.; Pardon, P.; Boyen, P. Rapid identification of 1 Mycoplasma bovis from bovine bronchoalveolar lavage fluid with MALDI-TOF MS after enrichment procedure. J. Clin. Microbiol. 2020, 58, e00004-20.
- Bell-Rogers, P.; Parker, L.; Cai, H.Y. Multi-locus sequence types of Mycoplasma bovis isolated from Ontario, Canada in the past three decades have a temporal distribution. J. Vet. Diagn. Invest. 2018, 30, 130–135.
- Register, K.B.; Lysnyansky, I.; Jelinski, M.D.; Boatwright, W.D.; Waldner, M.; Bayles, D.O.; Pilo, P.; Alt, D.P. Comparison of two multilocus sequence typing schemes for Mycoplasma bovis and revision of the PubMLST reference method. J. Clin. Microbiol. 2020, 58, e00283-20.
- Yair, Y.; Borovok, I.; Mikula, I.; Falk, R.; Fox, L.K.; Gophna, U.; Lysnyansky, I. Genomics-based epidemiology of bovine Mycoplasma bovis strains in Israel. BMC Genom. 2020, 21, 70.
- Parker, A.M.; Shukla, A.; House, J.K.; Hazelton, M.S.; Bosward, K.L.; Kokotovic, B.; Sheehy, P.A. Genetic characterization of Australian Mycoplasma bovis isolates through whole genome sequencing analysis. Vet. Microbiol. 2016, 196I, 118–125.
- Rasheed, M.A.; Qi, J.; Zhu, X.; Chenfei, H.; Menghwar, H.; Khan, F.A.; Guo, A. Comparative genomics of Mycoplasma bovis strains reveals that decreased virulence with increasing passages might correlate with potential virulence-related factors. Front. Cell. Infect. Microbiol. 2017, 7, 177.
- Maunsell, F.P.; Woolums, A.R.; Francoz, D.; Rosenbush, R.F.; Step, D.L.; Wilson, D.J.; Janzen, E.D. Mycoplasma bovis infections in cattle. ACVIM Consensus Statement. J. Vet. Intern. Med. 2011, 25, 772–783.
- Sachse, K.; Pfützner, H.; Hötzel, H.; Demuth, B.; Heller, M.; Berthold, E. Comparison of various diagnostic methods for the detection of Mycoplasma bovis. Rev. Sci. Tech.-Off. Int. Epiz. 1993, 12, 571–580.
- Hazelton, M.S.; Morton, J.M.; Parker, A.M.; Sheehy, P.A.; Bosward, K.L.; Malmo, J.; House, J.K. Whole dairy herd sampling to detect subclinical intramammary Mycoplasma bovis infection after clinical mastitis outbreaks. Vet. Microbiol. 2020, 244, 108662.
- Nielsen, P.K.; Petersen, M.B.; Nielsen, L.R.; Halasa, T.; Toft, N. Latent class analysis of bulk tank milk PCR and ELISA testing for herd level diagnosis of Mycoplasma bovis. Prev. Vet. Med. 2015, 121, 338–342.
- Gille, L.; Callens, J.; Supré, K.; Boyen, F.; Haesebrouck, F.; Van Driessche, L.; van Leenen, K.; Deprez, P.; Pardon, B. Use of breeding bull and absence of a calving pen as a risk factors for the presence of Mycoplasma bovis in dairy herds. J. Dairy Sci. 2018, 101, 8284–8290.
- Andersson, A.-M.; Aspán, A.; Wisselink, H.J.; Smid, B.; Ridley, A.; Pelkonen, S.; Autio, T.; Lauritsen, K.T.; Kensø, J.; Gaurivaud, P.; et al. A European inter-laboratory trial to evaluate the performance of three serological methods for diagnosis of Mycoplasma bovis infection in cattle using latent class analysis. BMC Vet. Res. 2019, 15, 369.
- Wawegama, N.K.; Browning, G.F.; Kanci, A.; Marenda, M.S.; Markham, F.F. Development of a Recombinant Protein-Based Enzyme-Linked Immunosorbent Assay for Diagnosis of Mycoplasma bovis Infection in Cattle. Clin. Vaccine Immunol. 2014, 21, 196–202.
- Vähänikkilä, N.; Pohjanvirta, T.; Haapala, V.; Simojoki, H.; Soveri, T.; Browning, G.F.; Pelkonen, S.; Wawegama, N.K.; Autio, T. Characterisation of the course of Mycoplasma bovis infection in naturally infected dairy herds. Vet. Microbiol. 2019, 231, 107–115.
- Petersen, M.B.; Wawegama, N.K.; Denwood, M.; Markham, P.F.; Browning, G.F.; Nielsen, L.R. Mycoplasma bovis antibody dynamics in naturally exposed dairy calves according to two diagnostic tests. BMC Vet. Res. 2018, 14, 258.
- Wisselink, H.J.; Smid, B.; Plater, J.; Ridley, A.; Andersson, A.-M.; Aspán, A.; Pohjanvirta, T.; Vähänikkilä, N.; Larsen, H.; Høgberg, J.; et al. A European interlaboratory trial to evaluate the performance of different PCR methods for Mycoplasma bovis diagnosis. BMC Vet. Res. 2019, 15, 86.
- Chazel, M.; Tardy, F.; Le Grand, D.; Calavas, D.; Poumarat, F. Mycoplasmoses of ruminants in France: Recent data from the national surveillance network. BMC Vet. Res. 2010, 6, 32.
- Mackie, D.P.; Finley, D.; Brice, N.; Ball, H.J. Mixed mycoplasma mastitis outbreak in a dairy herd. Vet. Rec. 2000, 147, 335–336.
- Jasper, D.E. Prevalence of mycoplasmal mastitis in the western states. Calif. Vet. 1980, 43, 24–26.
- McAuliffe, L.; Ellis, R.J.; Lawes, J.R.; Ayling, R.D.; Nicholas, R.A.J. 16S rDNA PCR and denaturing gradient gel electrophoresis; a single generic test for detecting and differentiating Mycoplasma species. J. Med. Microbiol. 2005, 54, 731–739.
- Conrad, C.C.; Daher, R.K.; Stanford, K.; Amoako, K.K.; Boissinot, M.; Bergeron, M.G.; Alexander, T.; Cook, S.; Ralston, B.; Zaheer, R.; et al. A Sensitive and Accurate Recombinase Polymerase Amplification Assay for Detection of the Primary Bacterial Pathogens Causing Bovine Respiratory Disease. Front. Vet. Sci. 2020, 7, 208.
- Loy, J.D.; Leger, L.; Workman, A.M.; Clawson, M.L.; Bulut, E.; Wang, B. Development of a multiplex real-time PCR assay using two thermocycling platforms for detection of major bacterial pathogens associated with bovine respiratory disease complex from clinical samples. J. Vet. Diagn. Invest. 2018, 30, 837–847.
- Pereyre, S.; Tardy, F.; Renaudin, H.; Cauvin, E.; Del Prá Netto Machado, L.; Tricot, A.; Benoit, F.; Treilles, M.; Bébéar, C. Identification and Subtyping of Clinically Relevant Human and Ruminant Mycoplasmas by Use of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry J. Clin. Microbiol. 2013, 51, 3314–3323.
More
