Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Vivi Li and Version 1 by Thevivanayagam Maharajan.
Cereals have evolved various tolerance mechanisms to cope with abiotic stress. Understanding the abiotic stress response mechanism of cereal crops at the molecular level offers a path to high-yielding and stress-tolerant cultivars to sustain food and nutritional security. In this regard, enormous progress has been made in the omics field in the areas of genomics, transcriptomics, and proteomics. Omics approaches generate a massive amount of data, and adequate advancements in computational tools have been achieved for effective analysis.
- abiotic stress
- functional genomics
- transcriptomics
- proteomics
- stress response
1. Introduction
The plant is a sessile organism that serves as the foundation for all living organisms on Earth and as a valuable resource for humans. All cereal crop species are members of the grass (Poaceae or Gramineae) family, the fourth largest family of flowering plants [1]. The major cereal crops, rice, maize, sorghum, finger millet, foxtail millet, wheat, barley, and other cereal crops, are the world’s primary food sources [2]. Food is necessary to provide energy for body growth, function, and defense. Our body’s basic requirements include proteins, minerals, carbohydrates, fiber, vitamins, and lipids, all essential nutrients. Among these requirements, carbohydrates, proteins, and fats which are needed in large quantities are called “macronutrients,” and the requirements of minerals and vitamins needed in small amounts are called “micronutrients” [3]. Cereals such as rice are enriched with carbohydrates; wheat is rich in carbohydrates and certain vitamins (B6); maize is rich in vitamins and micro elements, viz., P, Mg, Mn, Ca, Zn, Cu and Fe (depending on the varieties); millets are rich in fiber and proteins (https://data.nal.usda.gov/ (accessed on 16 October 2022)). These cereals are essential for the routine life of humans, and their production should be increased owing to the increasing population. However, abiotic stress is a global problem, limiting global crop production and reducing the yields of the plants by more than 50% in quality [4]. Abiotic stress conditions such as drought, high temperatures, salinity, mineral deficiency, and metal toxicity are usually experienced by plants in both natural and agricultural systems [5,6,7][5][6][7]. Among these stress factors, drought (low water availability), heat (extreme temperature), and salinity (high salt content) are the most important stresses, having a huge effect on the growth and productivity of the crops [8,9,10][8][9][10].
Furthermore, the world population is expected to reach nine billion by 2050, affecting average living standards. Food consumption also demands grain for livestock maintenance and agricultural land use, leading to the population’s food demand [8,11][8][11]. According to the FAO’s SOFI 2021 report (The State of Food Security and Nutrition in the World), the COVID-19 pandemic exposed agri-food systems to stress, resulting in increasing worldwide food instability and malnutrition [12]. There is an urgent need to increase crop productivity to meet the demand. Plants have developed sophisticated mechanisms to adapt to environmental changes and challenges that aid plant survival. The mechanism behind the environmental stress response in plants is undoubtedly more advanced and prominent than in animal cells [13]. Numerous fundamental reports are available on the molecular and cellular mechanisms behind abiotic stress adaptation in rice plants, which act as model plants for cereal crops. A plant’s response to drought, extreme temperature, and salinity depends on the regulation of genes (upregulation or downregulation). In this context, integrated omics research has been widely used to understand the plant’s biological networking and molecular mechanisms against various abiotic stresses. Despite tremendous progress in genomics, there is a need to study other omics levels, including transcriptomic and proteomic profiling, for a comprehensive understanding at the molecular level. Developing new crops that have improved resistance to various abiotic stress factors is essential. Plants are exposed to various abiotic stresses. Numerous stress-responsive genes are activated, which are involved in producing many proteins that help them activate and adjust the physiological and biochemical pathways in stress tolerance [14]. Therefore, integrating multi-omics data within the context of systems biology can provide more profound knowledge for future directions (molecular biology, genome editing, etc.) [15,16,17][15][16][17]. In this entreview, wey, researchers discussed the recent advancements in functional genomics, transcriptomic, and proteomic analyses to understand the adaptation and tolerance mechanisms enhancing drought, extreme temperature, and salinity in different cereal crops. A summary of transcriptomic, proteomic, and functional genomic approaches for crop improvement is presented (Figure 1).
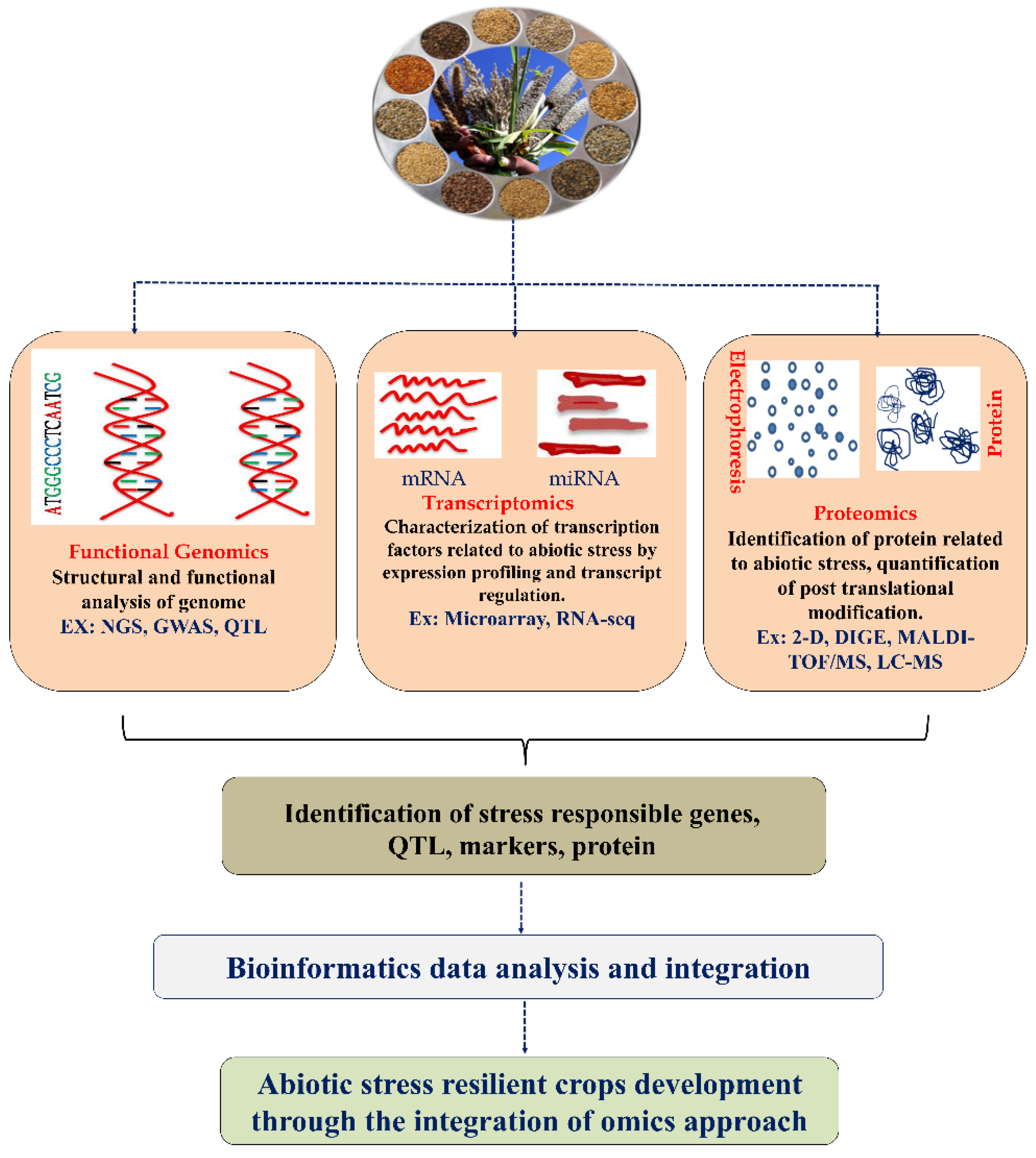
Figure 1. A schematic representation of omics approaches for identification of major regulator of stress response in the plant. QTL—quantitative trait locus; MAS—marker-assisted selection 2D gel electrophoresis; DIGE—differential gel electrophoresis; MALDI-TOF—matrix-assisted laser desorption/ionization time-of-flight; LC-MS/MS—liquid chromatography/mass spectrometry. Sources of the cereal image (https://agritech.tnau.ac.in/agriculture/millets_miracle_strategies.html (accessed on 17 October 2022)).
2. General Effects of Drought, Heat, and Salt Stress on Plant Growth and Development
Plants are multicellular, and their reactions to abiotic stresses are highly complicated and have passive-aggressive behavior. Depending on the environmental conditions, plants suffer from abiotic stresses that are either reversible or irreversible [14,17,18][14][17][18]. Besides that, how do plants react to environmental stress factors? Plant biologists and agronomists consider this to be an extremely severe problem due to the fact that it poses a significant risk to crop productivity. Plants have developed several biochemical, physiological, and metabolic responses to withstand environmental stress factors. Understanding plants’ molecular, cellular, physiological, and biochemical changes during abiotic stresses is crucial for better crop management [16,19,20][16][19][20]. Plants have primary and secondary stress response mechanisms to protect themselves from various abiotic stresses. The drought stress response involves the control of ion homeostasis (activation/inactivation of aquaporins), and water transport. Another general plant stress response is the synthesis of protective molecules or osmolytes, such as sugars, proline, polyalcohols, quaternary ammonium compounds, and so on, as well as different specific proteins such as heat shock proteins, LEA proteins, osmotic, etc., [16,19,20][16][19][20]. In the secondary stress response, plants generate “reactive oxygen species” (ROS), which includes H2O2 peroxidation and lipid peroxidation (MDA content was increased). Reduced glutathione, superoxide dismutase, catalase, ascorbate peroxidase, glutathione peroxidase, and other antioxidant enzymes are activated when ROS accumulates in the cell cytoplasm [21]. The general effects of drought, heat and salt stress on plant growth and development have been shown (Figure 2).
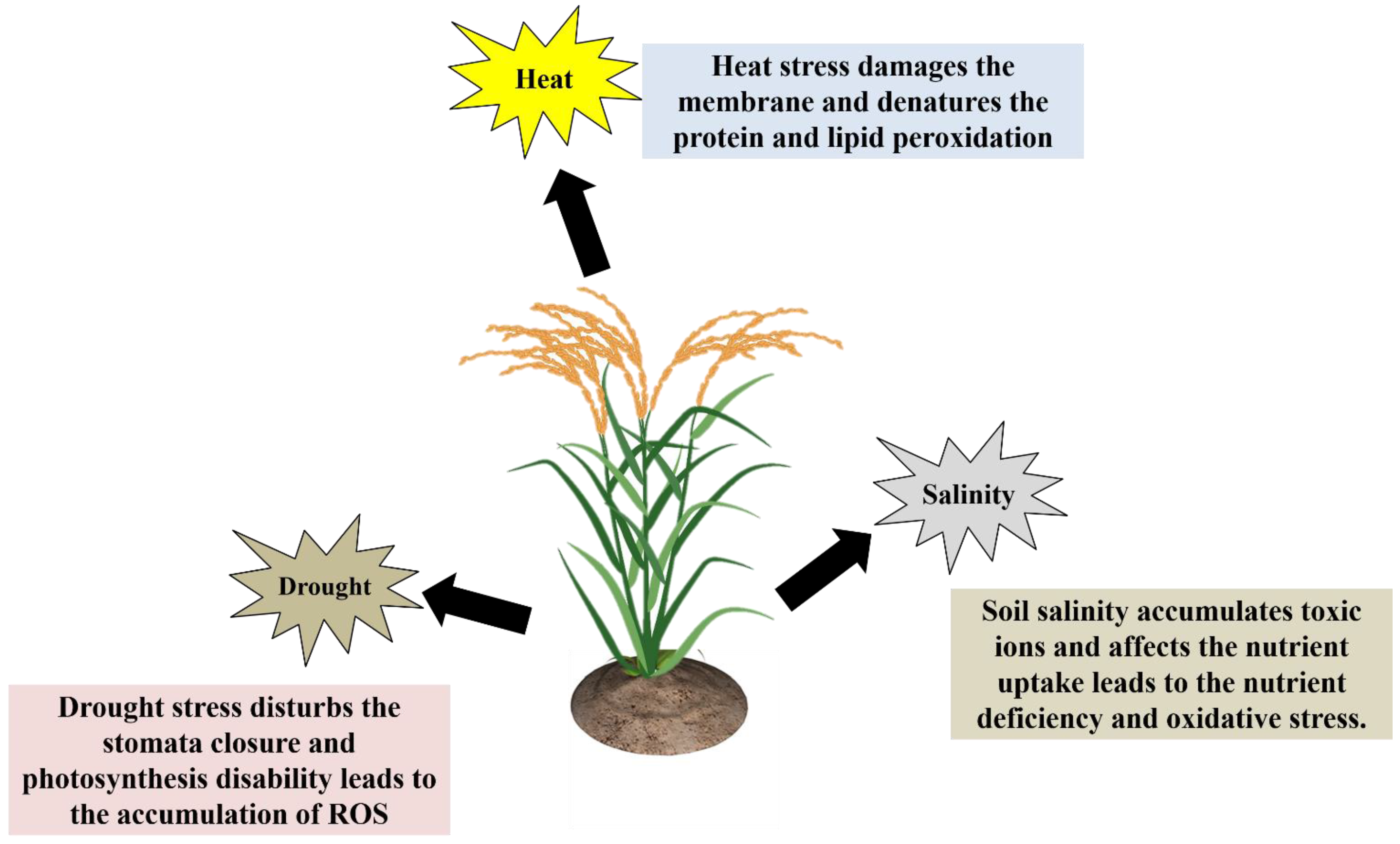
Figure 2.
How do plants react to environmental stress factors?
2.1. Drought Stress
The stress response of plants varies with each species, determined by the growth stage and environmental factors [22]. Drought stress reduces the yield of maize (63–87%) [23], wheat (57%) [24], and rice (53–92%) [25]. The underlying impact of drought is poor germination, damaged seedling development, decreased root and shoot dry weight, hypocotyl length, and vegetative development and these factors have been accounted for in imperative nutrition crops including rice, wheat, sorghum, foxtail millet, and finger millet, etc., [26,27,28,29,30][26][27][28][29][30]. Water and nutrient associations are important factors for the growth and development of plants. Stomatal conductance, leaf water potential, transpiration rate, and water relations are activated. Drought stress highly affects the above factors and the nutrient relationships such as N, Si, Mg, and Ca [31]. Furthermore, drought stress decreases the soil moisture and leads to a reduction in root growth, which is deeper and thicker. In this context, roots are the first organ that senses water availability and sends signals to the aerial organs via root-to-shoot xylem channels to decrease the turgidity of guard cells and close the stomatal aperture to reduce water loss [22,32][22][32]. Drought-responsive genes were divided into two categories: functional and regulatory genes. Synthase genes, aquaporin genes, protective metabolites (sucrose, proline, and betaine), and proteins (LEA protein, molecular chaperone) are all involved in cell defense against environmental stress. Stress-related transcription factor genes, protein kinase genes, phospholipid metabolism-related genes, and protein phosphatase genes are regulatory gene products that indirectly protect plant cells from abiotic stress [16,33,34,35][16][33][34][35].
2.2. Temperature Stress (Heat)
Temperature is a major factor affecting plants’ distribution, growth, and development. Plant morphological and physiological processes are affected by the temperature conditions under which the plant species grow, because each species grows in a specific temperature range (i.e., 25–30 °C for maximum growth) [36]. Elevated temperatures cause adverse effects such as the burning of shoots and leaves, leading to leaf senescence and growth inhibition [22,37][22][37]. Under this condition, plants try to balance their tissue water and moisture content to protect themselves from high temperatures. It can also reduce the germination, development of spikes, number of florets, and net assimilation rate in sorghum, rice, and maize [22,37][22][37]. Heat stress independently alters the physiology and metabolism of plants. However, its effect becomes enhanced when combined with other abiotic stresses such as drought and salt stress [22,38][22][38]. Heat stress in the reproductive stage causes a major reduction in the yield of crops due to the poor photosynthetic process. Extreme temperature stress leads to the deactivation of various enzymes involved in photosynthesis. For example, PSII enzymes are an important factor in photosynthesis; high temperatures influence the activity of PSII. Under high temperatures, components of PSII were damaged in wheat and barley [38,39][38][39]. Although high-temperature stress reduces the enzyme activities of adenosine diphosphate-glucose pyrophosphorylase, sucrose phosphate synthase, and invertase, it affects the starch and sucrose synthesis of plants [40,41][40][41].
2.3. Salinity Stress
Salinity is an abiotic stress that severely affects plant growth, development, and crop production. The response of plants under salinity stress can be described in two phases: the initial phase, an ion-independent response, which takes minutes to days to cause toxicity, affects stomatal closure and inhibits cell expansion, particularly in the shoot [22,42][22][42]. The second phase, the ion-dependent response, which takes days or weeks to build up cytotoxicity, causes premature senescence of leaves, reducing yield or even causing death [42,43,44][42][43][44]. Moreover, salt may affect plant growth indirectly by decreasing the rate of photosynthesis and stomatal conductance [45,46][45][46]. Stomata are the main structures responsible for gas exchange control. Salt stress reduces stomatal conductance by affecting their opening, size, and density [47]. Consequently, transpiration (i.e., water loss) and photosynthesis (CO2 uptake) rates are also reduced [48,49][48][49]. Increased Na+ ion is sensed by plants, leading to immediate closure of stomata and inhibition of leaf expansion within minutes of exposure. Later, due to excessive ion concentration, premature senescence of leaves and a reduction in yield occur in plants [47]. Compared to other cereals, rice is the most salt-sensitive crop. Chlorophyll and carotenoid contents in rice leaves were significantly decreased after the introduction of salt stress [50]. An excess amount of salt adversely affects the metabolic activities of plants, including cell wall damage, accumulation of electron-dense proteinaceous particles, plasmolysis, cytoplasmic lysis, and damage to ER. In addition, it accumulates citrate, malate, and inositol in leaf blades within one day of salt treatment [51]. Sorghum is a moderate, salt-tolerant agronomic crop. Swami et al. [52] reported that sorghum plants under salt-stress conditions reduced the growth of leaves and chlorosis. In sweet sorghum, salinity decreased germination percentage and increased germination duration [53,54][53][54].
2.4. Oxidative Damage and Antioxidant Enzyme Defense System in Plants under Abiotic Stress
Abiotic stresses affect plant growth, development, and productivity by degrading cellular metabolism and increasing reactive oxygen species (ROS) generation. During abiotic stress, photorespiration, the photosynthetic system, and mitochondrial respiration pathways contribute to the generation of ROS [55,56][55][56]. It has been proven that ROS are generated in different cellular compartments such as mitochondria, chloroplasts, peroxisomes, cytoplasm, and the extracellular region [17,57,58,59][17][57][58][59]. The increased production of ROS during stress can be damaging to cells. Still, reactive oxygen intermediate (ROI) acts as a signal to activate stress-response and defense pathways [60]. Thus, ROS can be considered a cellular indicator and secondary messenger in the stress-response signal transduction pathway. Approximately 1–2% of total molecular oxygen consumption results in the production of ROS in normal conditions. The stressed plant cell generates toxic ROS such as hydrogen peroxide (H2O2), singlet oxygen (1O2), alkoxyl (RO•), peroxyl (ROO•), hydroxyl radical (OH•), and superoxide (O2•−)[17,57,59,61,62][17][57][59][61][62]. Increased ROS levels can potentially cause peroxidation of lipids, denaturation of proteins, mutation of DNA, and cellular oxidative damage [59,63][59][63]. The equilibrium between ROS production and elimination at the intracellular level must be regulated to overcome ROS induced damage, and maintain the growth, metabolism, development, and overall productivity of the crops. Plants cells produce a complex of enzymatic and nonenzymatic antioxidants to maintain ROS homeostasis. The enzymatic antioxidant system includes several antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), glutathione reductase (GR), peroxiredoxins (Prxs), and enzymes of the ascorbate-glutathione (AsAGSH) cycle, such as ascorbate peroxidase (APX), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR). Nonenzymatic antioxidants such as glutathione (GSH), ascorbate (AsA), tocopherol, carotenoids, and phenolic compounds are also involved in the removal of ROS [64]. According to several reports, antioxidant enzyme activity is involved in the stress tolerance of plants. Alterations in SOD, CAT, APX, and GR activity were observed in various cereal crops under abiotic stress [65].
Superoxide dismutase is the most effective intracellular metalloenzyme that serves as the first line of defense against ROS-mediated oxidative stress. According to the metal cofactors, SODs are classified into three types: Cu/Zn-SOD (copper/zinc cofactor), Mn-SOD (manganese cofactor), and Fe-SOD (iron cofactor) [66,67,68,69][66][67][68][69]. SOD catalyzes the dismutation of the superoxide radicals (O2.−) into hydrogen peroxide (H2O2) and oxygen (O2) and decreases the formation of hydroxyl radical (OH•) formation via the metal-catalyzed Haber–Weiss-type reaction. The dismutation rate is 10,000-fold higher than spontaneous dismutation [59]. Under stressful environments, upregulation of SOD has been associated with plant survival and mitigation of oxidative damage. A significant increase in SOD activity under drought stress has been observed in many plant species, viz., rice [70], wheat [71], sorghum, and sunflower [72].
Catalase is a tetrameric heme-containing enzyme involved in degrading H2O2 into H2O and O2. Under normal environmental conditions, catalase scavenges H2O2 produced during photorespiratory oxidation, mitochondrial electron transport, and β-oxidation of the fatty acids. Catalase enzyme is found in the mitochondria, peroxisomes, and cytoplasm of higher plants [73]. SOD detoxifies superoxide radicals (O2.−) into hydrogen peroxide (H2O2) and oxygen (O2), then then the H2O2 can be eliminated by CAT.
In plant cells, AsA-dependent APX (EC 1.11.1.1) occurs in different isoforms (cytosolic APX (cAPX), mitochondrial APX (mtAPX), chloroplastic APX (chlAPX) and peroxisomal/glyoxysomal APX (mAPX). APX is the only enzyme capable of scavenging H2O2 in the chloroplast since CAT is not present, which participate in the AsA-GSH cycle producing monodehydroascorbate (MDHA) [74].
GR is a flavoprotein oxidoreductase involved in the defense system by sustaining the status of glutathione (GSH), a disulfide reductant, which protects thiol groups of enzymes, regenerates ascorbate, and reacts with singlet oxygen (1O2) and hydroxyl radical (OH•). GSH and GR play a key role in determining the tolerance of a plant under various stresses. This might be due to maintaining a high ratio of NADP+/NADPH, therefore ensuring the availability of NADP+ for accepting electrons from the photosynthetic electron transport chain and facilitating the regeneration of oxidized ascorbate [17,60,65][17][60][65].
3. Functional Genomic Approaches to Identify Stress-Responsive Genes in Cereals
The genomics strategies help develop climate-resilient crop varieties to ensure food security by helping conserve genomic resources. The discovery of genomic variations and genes related to climate adaptation found in wild relatives of crop plants through whole-genome sequencing is an indicator of the development of environmentally-adapted crops [75,76][75][76]. Genomics and systems biology approaches toward the discovery of stress-tolerant traits have been made possible by the availability of molecular markers, QTL mapping, genetic mapping, and comparative genomics, the complex relationship of plants and environmental factors, and expression quantitative trait loci (eQTL). These techniques can provide a practical approach to identifying candidate genes involved in abiotic stress tolerance.
3.1. Genome Sequencing
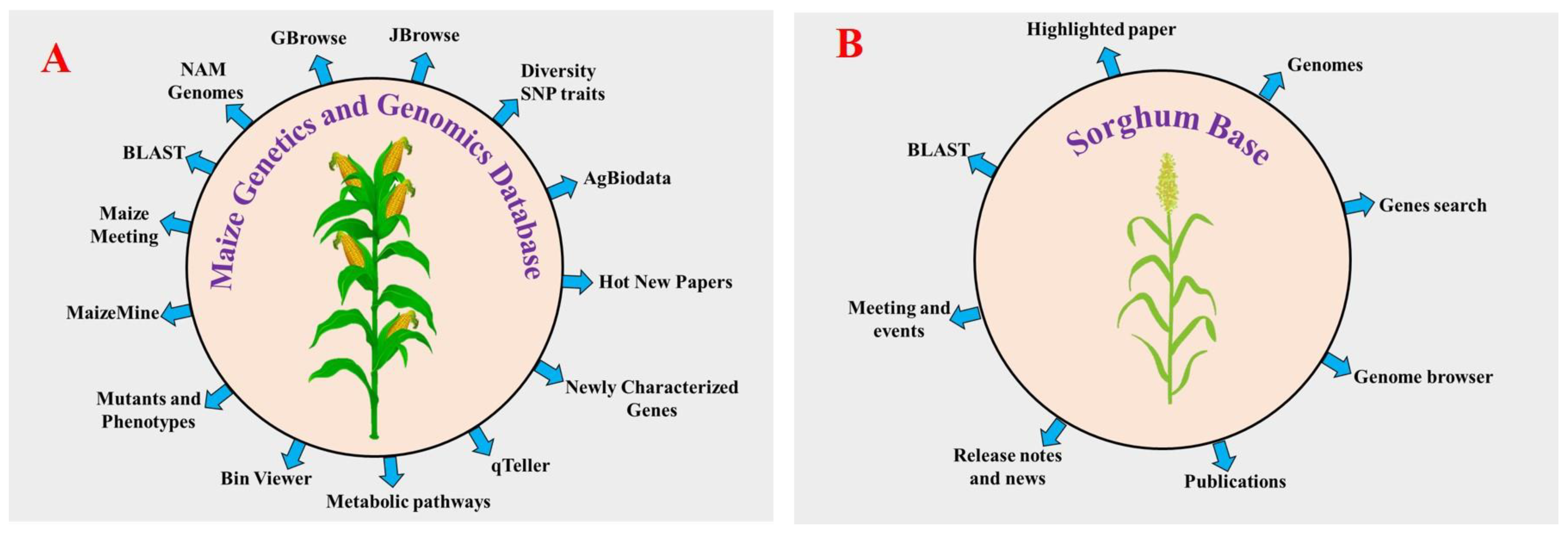
Rice was the first cereal to be sequenced, which paved the way for NGS characterization of more complex cereals [77,78,79][77][78][79]. Afterward, most commonly available cereals had their genomes sequenced at the whole genome level, including maize [80], sorghum [81], foxtail millet [82[82][83],83], wheat [84], barley [85], finger millet [86], pearl millet, kodo millet, barnyard millet and green foxtail. Over the past years, advancements have been made in DNA sequencing technology, facilitating the generation of a large amount of sequencing data within a short time in a cost-effective manner compared with the first-generation sequencing methods (Sanger-sequencing). Next-generation sequencing (NGS) technologies have high-throughput sequencing with prominent next-generation sequencing (NGS) platforms, including the Illumina/Solexa AB SOLiD Genome analyzer (https://www.illumina.com/ (accessed on 18 January 2022)) and Roche 454 GS FLX Titanium (www.454.com (accessed on 20 January 2022)). These technologies increase the eagerness for genome sequencing. First- and second-generation sequencing technologies have some disadvantages, so the researchers developed third-generation sequencing technologies to overcome certain drawbacks of first- and second-generation sequencing methods. In whole-genome sequencing of plants, single molecule sensor (HeliScope™) [87], single molecule real time sequencer (SMRT™) [88[88][89],89], single molecule real time sequencer (RNAP™) [90], and Nanopore DNA sequencers are used. They illustrate detailed genomic features including coding and noncoding genes, GC content, and repetitive and regulatory sequences that can be used to understand the functional roles of plant genes [91,92][91][92]. The NGS technology has opened the door to studying plant genomics to understand the abiotic stress response and to also produce improved crop varieties against abiotic stress. The study of plant species with larger genome sizes is more complicated due to the presence of repetitive elements in their genomes. These complex genomes are one of the most challenging problems for sequence assembly. For example, the genome size of the monocot plant wheat (hexaploid) is highly complex, consisting of 17 Gb, compared with the 4.79 Gb barley genome (diploid) [93,94][93][94]. Among the cereals, genome databases are available for rice and maize. Four other data genome databases are available for rice such as Rice Genome Annotation Project (RGAP) (http://rice.uga.edu/ (accessed on 25 January 2022)), International Rice Informatics Consortium (IRRI) (http://iric.irri.org/resources/rice-databases (accessed on 2 February 2022)), O. sativa genome database (OsGDB) (https://www.plantgdb.org/OsGDB/ (accessed on 12 February 2022)) and Rice RelativesGD (http://ibi.zju.edu.cn/ricerelativesgd/ (accessed on 17 February 2022)) [95,96,97][95][96][97]. All these databases contain annotated genomes of rice, and molecular resources (cDNA full length, gene, EST, markers, and expression data). The first genome sequence of maize was released in 2009 by Schnable et al. [80]. Researchers released several genome assemblies of maize from different countries afterwards. The maize genetics and genomics database (MaizeGDB) is the community database and global web source for maize [98,99,100][98][99][100]. More the 40 genome assemblies of maize inbred lines are available in addition to the B73 reference genome assembly. To date, MaizeGDB contains the five B73 reference genome assemblies of maize. The current B73 assembly version, Zm-B73-REFERENCE-NAM-5.0 (also known as RefGen_v5), released in January 2020, was sequenced and assembled along with a set of 25 inbred known as the Nested Associated Mapping (NAM) founder lines by the NAM Consortium using PacBio long reads and mate-pair strategy. The RefGen_v5 assembly sequence includes all 10 chromosomes. The first three assemblies (RefGen_V1, V2, and V3) were based on a bacterial artificial chromosome (BAC) sequencing strategy. The RefGen_V4 assembly used a new approach that relied on PacBio SMRT sequencing at Cold Spring Harbor to a depth of 60x coverage with scaffolds created via the assistance of whole genome restriction mapping (aka Optical Mapping). Maize-GDB also contains 38 additional reference genome assemblies, such as eight inbred lines (A188, B104, CMC247, Mo17, PH207, and W22), four European flints (DK105, EP1, F7, and PE0075), 25 NAM population (B97, CML52, 69, 103, 228, 247, 277, 322, 333, HP301, Il14H, Ki3, Ki11, Ky21, M37W, M162W, Mo18W and Ms71) and one teosinte (PI566673). Around 60 % of genes are found in all the NAM lines. Researchers have extensively used NAM populations to study maize flowering time, leaf structure, disease resistance, and other important agronomic traits. MaizeGDB hosts a wide range of data, including genome metadata, microRNAs, QTLs, SNPs, genome assemblies, genetic maps, ESTs, pathway da-ta, microarray data, RNA-seq, proteins, gene models, and transcripts. Several tools such as qTeller, maizemine, metabolic pathways, maize meeting, bin viewer, newly characterized genes, diversity SNP traits, and AgBioData have been implemented to improve the accessibility and visualization of data (Figure 3) [101]. Among these, qTeller is a comparative RNA-seq expression platform that helps to compare expression between two genes visually. Maizemine is a data mining respiratory for the MaizeGDB [102]. It enables researchers to create and export customized annotated datasets that can be combined with their research data for use in downstream analyses. Using the maize meeting tool, researchers can find past, present, and future information about maize genetics meetings. These recent updates at MaizeGDB will serve as a template for other databases to manage large-scale pan-genomes of any species. More than 15 genome assemblies are currently available at the SorghumBase database (https://sorghumbase.org/ (accessed on 18 October 2022)). As with rice, sorghum and maize, the development of genomic databases for other cereals will enable researchers to gather information about all molecular data.

Figure 3. Graphical illustration of the maize genetics and genomics database (MaizeGDB) and sorghum base. Various tools have been implemented in the MaizeGDB (A) (https://www.maizegdb.org (accessed on 10 October 2022)) and sorghum base (B) (https://www.sorghumbase.org (accessed on 18 October 2022)), both of which help to collect all molecular data from the database.
Among the small millets, foxtail millet genome sequence has been sequenced by two research institutes (US Department of Energy Joint Genome Institute and Beijing Genomics Institute) [83,103][83][103]. The genome size of foxtail millet is around 515 Mb. A total of 38,801 genes were predicted (30,579 genes annotated and 8220 genes unannotated) from the genome sequence of foxtail millet. Bennetzen et al. [83] predicted more than 25,000 protein-encoding genes and 63,286 expressed sequence tags (ESTs) from their foxtail millet genome sequence. Like foxtail millet, two research groups released finger millet’s draft genome sequence in 2017 and 2018 [86,104][86][104]. The Hittalmani et al. [86] finger millet draft genome consisted of 525,759 scaffolds (>200 bp) with N50 length of 23.73 Kb, and the average scaffold length of 2275 bp. They have predicted 78,647 non-transposable elements and 6596 transposable elements-related genes from their draft genome sequences of finger millet using a de novo gene prediction method using Augustus. Furthermore, they have also predicted drought stress-responsive genes (2866), calcium transport and accumulation genes (330), and C4 pathway genes (146) from the same draft genome sequence of finger millet. The released draft genome sequences of finger millet are publicly available at the National Center for Biotechnology Information (NCBI) (Bio-Sample numbers: SAMD00076255 and SAMN04849255). The School of Plant Sciences, Ecology and Evolutionary Biology, Arizona Genomics Institute recently released a thoroughly annotated genome sequence (version 1.0 assembly) at the Phytozome database (https://phytozome-next.jgi.doe.gov/info/Ecoracana_v1_1 (accessed on 4 October 2022)). The genome assembly was generated by a MECAT assembler [105] and subsequently polished using QUIVER. The released genome assembly (version 1.0) contains a 1110.3 Mb sequence, consisting of 674 contigs. In 2018, the China Agricultural University submitted the shotgun genome sequence of proso millet (923 Mb) at NCBI (bio-sample and bio-project numbers are SAMN08335224 and PRJNA429322, respectively) [106]. Around 55,930, 339, 1420, 1640 and 2302 protein-coding genes, microRNAs, transfer RNAs, ribosomal RNAs and small nuclear RNAs were identified from the draft genome sequences of proso millet, respectively. The draft genome sequence for barnyard millet was generated by [107]. The genome size of barnyard millet was estimated to be 1.27 Gb with a scaffold N50 length of 1.8 Mb. About 917 cytochrome P450 monooxygenase, 277 glutathione S-transferase, 4945 differentially expressed genes, 108,771 protein-coding genes, 785 microRNAs and other non-coding RNAs were predicted from the draft genome sequence of barnyard millet. The International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, India, has released the draft genome sequence of pearl millet (~1.79 Gb) [108]. They used whole genome shotgun and bacterial artificial chromosome sequencing techniques to assemble the pearl millet genome. About 27,893 (72.30%) genes were annotated and 10,686 (27.70%) genes unannotated. The annotated and drafted genome sequences of all cereals will help to identify the genes and markers and will help to improve the growth and production of cereals under both biotic and abiotic stresses. With the enormous amount of data obtained from these sequencing methods, the major question is how to analyze and utilize this information. This has prompted scientists to develop bioinformatics tools capable of extracting biologically meaningful information from a large amount of data. The genome assemblers are TIGR assembler [109], CAP3 [102], string graph assembly [110], Newbler [111], SSAKE [112], VCAKE [113], SHARCGS [114], ALLPATHS-LG [115], Edena [116], Velvet [117], CABOG [118], ABySS [119], Genomic Analysis Toolkit [120], and SOAPdenovo2 [121]. They have been developed to reconstruct the whole genome sequence by aligning and merging sequence reads generated from the current genome sequencing technologies. CAP3 is the third generation of contig assembly programs. It helps to compute the assembly of large genomes such as mice, humans and maize [122]. CAP3 has been successfully used for the EST assembly of several plant species including, rice, wheat, and maize [123,124][123][124]. The quantitative trait loci sequencing (QTL-seq) strategies can be used to conquer the challenges of complex genomes. They can be used to detect QTL in large populations [125].
3.2. Molecular Markers
The availability of genome sequence data has provided new potential resources for cereal crop improvement. Still, the genome sequence’s large size and non-coding parts were bottlenecks associated with the genetic information. It is not easy to sequence and obtain information about the genes from proteomics. Expression sequence tags (EST) were developed to overcome the large and non-coding genome problems. With this method, a cDNA library can be produced from different tissues at different developmental stages and under different stress conditions, enabling the identification of transcripts related to tissues or stress-specific conditions. It is easy to obtain complicated and targeted plant species sequencing under abiotic stress conditions. It also provides the mRNA with a sufficient expression profile of specific sequences from cDNA under stressed conditions. EST data were generated and made available (http://www.ncbi.nlm.nih.gov/dbEST (accessed on 5 March 2022)). The NCBI-EST database contains approximately 449,101 ESTs for drought, 312,353 ESTs for salinity, 103,898 ESTs for low temperature, and 252,595 ESTs for high temperature [127,128][126][127]. PlantGDB (http://www.plantgdb.org/ (accessed on 5 October 2022)) provides genome browsers to display current gene structure models and transcript evidence from spliced alignments of EST and cDNA sequences. CerealESTdb comprises ESTs from four major cereal crops, namely rice (Oryza sativa L.), wheat (Triticum aestivum L.), sorghum (Sorghum bicolour L.), and maize (Zea mays L.), under various abiotic stress, viz., salt, drought, heat, cold and ABA. This database consists of 55,826 assembled EST sequences, 51,791 predicted genes models, and their 254,609 gene ontology terms including extensive information on 1746 associated metabolic pathways. The CerealESTdb is publicly available with the URL http://cabgrid.res.in/CerealESTDb (accessed on 5 October 2022) [129][128].
Molecular markers are the most important tools to develop and improve selection efficiency in identifying novel agronomic traits. Molecular markers are used for phylogeny and evolution studies, analysis of exotic germplasm diversity, cultivar genotyping, biotic and abiotic stress resistance, etc. [130][129]. A large number of molecular markers, such as SSRs (simple sequence repeats) and SNPs (single-nucleotide polymorphisms), have provided new strategies for germplasm characterization and genetic improvement, including breeding for resistance to abiotic stresses [131][130]. SSRs (simple sequence repeats) are 1–6 nucleotides contain multi-allelic and co-dominant at a single locus. The tandem arrays of SSR motifs mutate at the rate of 10–7 to 10–3 mutations per locus/generation [132][131]. Therefore, the number of repeat units in an individual genotype leads to high polymorphic and genotypic variation, making these sites useful for genetic analysis and breeding. A total of more than 20,000 SSR primers for rice have been developed [133][132]. McCouch et al. [134][133] developed 2240 di, tri, and tetra nucleotide markers and experimentally validated them. Among these, 56 SSR markers of O. sativa match with the BAC clone of the O. sativa genome. In rice, QTL-linked SSR markers generated 99 polymorphic alleles in 142 rice genotypes [135][134]. Based on the PIC (polymorphism information content) value (0.991), marker RM8094 proved to be an appropriate marker to distinguish a salt-tolerant rice variety from a salt-sensitive variety [135][134]. Similarly, RM104 is a marker to differentiate drought tolerance in rice. It is evident that the genomic region RM212–RM302–RM3825 on chromosome 1 is connected to drought resistance traits and can be beneficial in marker-assisted breeding for drought resistance in rice [136][135].
In recent years, SNP-based diversity analysis has been used in several plants [131,137][130][136]. It can dominate other molecular markers because of their irregular occurrence and minimal level of mutation [138][137]. The NCBI SNP network databases (https://www.ncbi.nlm.nih.gov/projects/SNP/ (accessed on 10 March 2022)) allow uresearchers to identify functional SNPs. CerealsDB (https://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/indexNEW.php (accessed on 5 October 2022)) was created by members of the Functional Genomics Group at the University of Bristol. It provides information about the SNP in the genomes of bread wheat (Triticum aestivum) and its relatives [139][138]. Various software is readily available to visualize, analyze, and identify SNPs, including SNPsniffer, SeqMan genome analyzer, Atlas-SNP, ssahaSNP, PanGEA, CLCbio, MAQ, and NextGENe [91,140][91][139]. SNPs have been used to identify QTLs and ESTs associated with drought, salinity, and heat stress. In rice, the regulatory region of ERF3 employed to detect functional SNPs in drought stress tolerance revealed 31 SNPs and short InDels [141][140]. TaDREB1 is an important transcription factor present in wheat plants, which is involved in drought stress. Chen et al. [142][141] identified 271 SNPs and 14 InDels of TaDREB1 transcription factor associated with drought stress. SNP marker analysis between the heat-tolerant (K7903) and heat-susceptible (RAJ4014) genotypes of heat shock protein HSP16.9 revealed the SNP (A/G) at the thirty-first amino acid position, resulting in a point mutation from Asp to Asn [143][142]. In Zea mays, association mapping with candidate genes leads to identifying SNPs associated with modifying abscisic acid levels in floral tissue during drought conditions [144][143]. Zea mays cultivars from Serbia and Bulgaria were used to identify the SNPs of a MYBE1 transcription factor. Only the Ser/Thr-rich region of MYBE1 is affected by SNP mutation, not the conserved R1 domain. The detected SNP mutations are associated with genes related to drought stress tolerance in maize [145][144]. The SNP linkage map of 50 barley genotypes in response to terminal drought stress during plant growth and development and the expression patterns of drought-regulated genes resulted in the identification of 17 starch synthesis/degradation genes [146,147,148][145][146][147].
3.3. Genome-Wide Association Studies (GWAS)
GWAS is a powerful tool because it can determine natural variations in all the recombination events that occur in the evolutionary processes of a wide range of organisms [170,171][148][149]. The main objective of the GWAS is to identify SNPs [172][150]. WGAS exploits the databases that contain reference plant genome sequences and genetic maps to analyse whole samples to find genetic variations based on complex traits such as growth rate, flowering time, and yield. They are the major focus of crops to improve the quality and understand the adaptation of plants. This sentudry revealed DNA samples of tolerant and susceptible plant varieties on a chip and they were scanned through an automated scanning machine. It can search the genetic variations for a selected marker such as SNPs if the variations associated with traits are found. One of the most important factors in GWAS is data quality. PLINK, a freely available tool set, can be used. It contains two data sets: one consists of individuals and their genotypes, and the other contains information on the genetic markers [172][150]. PLINK provides Principal Component Analysis (PCA) for population stratification and Bonferroni correction to control the false discovery rate. SNPs could be used in a PRS (Polygenic Risk Prediction Analyses) analysis. Genotypes with a p value > 5% missing rate and a p value < 1% allele frequency create problems for further analysis and thus are eliminated from the study [91]. GWASs were completed successfully on food crops such as rice, maize, sorghum, and foxtail millet. Huang et al. [173][151] identified approximately 3.6 million SNPs in 517 rice land races by GWAS. GWAS of 950 accessions of cultivar led to the identification of loci associated with flowering time and grain yield. They were genotyped into the rice land races by the sequencing-by-synthesis method. GWAS of S. bicolor was performed on 971 accessions, yielding approximately 265,000 SNPs. Loci were associated with height and maturity [174][152]. Using GWAS, Jia et al. [175][153] discovered 2.58 million SNPs in 916 foxtail millet (Setaria italica) varieties. Thise study grouped 916 foxtail millet varieties into five categories, and 512 loci are related to 47 agronomic traits. GWAS will allow comparative studies of adaptive genetic variation among species that have potentially evolved in parallel under selective stress. A better understanding of the adaptive responses of plants under stressed conditions might be obtained by comparing the genetic structures of adaptive traits among species.
References
- Salse, J.; Feuillet, C. Comparative Genomics of Cereals. Genom.-Assist. Crop Improv. 2007, 177–205.
- Lata, C.; Shivhare, R. Engineering Cereal Crops for Enhanced Abiotic Stress Tolerance. Proc. Indian Natl. Sci. Acad. 2021, 87, 63–83.
- TAŞĞIN, E. Macronutrients and Micronutrients in Nutrition. Int. J. Innov. Res. Rev. 2017, 1, 10–15.
- Ahmad, P.; Abdel Latef, A.A.H.; Rasool, S.; Akram, N.A.; Ashraf, M.; Gucel, S. Role of Proteomics in Crop Stress Tolerance. Front. Plant Sci. 2016, 7, 1336.
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) Gene Family in Rapeseed (Brassica Napus L.): Genome-Wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Int. J. Mol. Sci. 2021, 22, 4281.
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-Morphological Traits. Front. Plant Sci. 2017, 8, 537.
- Rani, S.; Kumar, P.; Suneja, P. Biotechnological Interventions for Inducing Abiotic Stress Tolerance in Crops. Plant Gene 2021, 27, 100315.
- Giordano, M.; Petropoulos, S.A.; Rouphael, Y. Response and Defence Mechanisms of Vegetable Crops against Drought, Heat and Salinity Stress. Agriculture 2021, 11, 463.
- Roy, S.J.; Tucker, E.J.; Tester, M. Genetic Analysis of Abiotic Stress Tolerance in Crops. Curr. Opin. Plant Biol. 2011, 14, 232–239.
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122.
- Comas, L.H.; Trout, T.J.; DeJonge, K.C.; Zhang, H.; Gleason, S.M. Water Productivity under Strategic Growth Stage-Based Deficit Irrigation in Maize. Agric. Water Manag. 2019, 212, 433–440.
- World Health Organization. The State of Food Security and Nutrition in the World 2021: Transforming Food Systems for Food Security, Improved Nutrition and Affordable Healthy Diets for All; Food & Agriculture Org.: Rome, Italy, 2021; Volume 2021, ISBN 925134325X.
- Wang, H.; Qin, F. Genome-Wide Association Study Reveals Natural Variations Contributing to Drought Resistance in Crops. Front. Plant Sci. 2017, 8, 1110.
- Pandian, S.; Rakkammal, K.; Rency, A.S.; Muthuramalingam, P.; Pandian, S.K.; Ramesh, M. Abiotic Stress and Applications of Omics Approaches to Develop Stress Tolerance in Agronomic Crops. In Agronomic Crops; Springer: Singapore, 2020; pp. 557–578.
- Zhan, X.; Lu, Y.; Zhu, J.; Botella, J.R. Genome Editing for Plant Research and Crop Improvement. J. Integr. Plant Biol. 2021, 63, 3–33.
- Paes de Melo, B.; de Carpinetti, P.A.; Fraga, O.T.; Rodrigues-Silva, P.L.; Fioresi, V.S.; de Camargos, L.F.; da Silva Ferreira, M.F. Abiotic Stresses in Plants and Their Markers: A Practice View of Plant Stress Responses and Programmed Cell Death Mechanisms. Plants 2022, 11, 1100.
- Yaqoob, U.; Jan, N.; Raman, P.V.; Siddique, K.H.M.; John, R. Crosstalk between Brassinosteroid Signaling, ROS Signaling and Phenylpropanoid Pathway during Abiotic Stress in Plants: Does It Exist? Plant Stress 2022, 4, 100075.
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of Abiotic Stress on Plants: A Systems Biology Perspective. BMC Plant Biol. 2011, 11, 1–14.
- Pandian, S.; Rakkammal, K.; Rathinapriya, P.; Rency, A.S.; Satish, L.; Ramesh, M. Physiological and Biochemical Changes in Sorghum under Combined Heavy Metal Stress: An Adaptive Defence against Oxidative Stress. Biocatal. Agric. Biotechnol. 2020, 29, 101830.
- Rathinapriya, P.; Pandian, S.; Rakkammal, K.; Balasangeetha, M.; Alexpandi, R.; Satish, L.; Rameshkumar, R.; Ramesh, M. The Protective Effects of Polyamines on Salinity Stress Tolerance in Foxtail Millet (Setaria Italica L.), an Important C4 Model Crop. Physiol. Mol. Biol. Plants 2020, 26, 1815–1829.
- He, M.; He, C.-Q.; Ding, N.-Z. Abiotic Stresses: General Defenses of Land Plants and Chances for Engineering Multistress Tolerance. Front. Plant Sci. 2018, 9, 1771.
- Chaudhry, S.; Sidhu, G.P.S. Climate Change Regulated Abiotic Stress Mechanisms in Plants: A Comprehensive Review. Plant Cell Rep. 2021, 41, 1–31.
- Kamara, A.Y.; Menkir, A.; Badu-Apraku, B.; Ibikunle, O. The Influence of Water Deficit on Growth, Yield and Yield Components of Some Maize Genotypes. J. Agric. Sci. 2003, 141, 43–50.
- Balla, K.; Rakszegi, M.; Li, Z.; Bekes, F.; Bencze, S.; Veisz, O. Quality of Winter Wheat in Relation to Heat and Drought Shock after Anthesis. Czech J. Food Sci. 2011, 29, 117–128.
- Lafitte, H.R.; Yongsheng, G.; Yan, S.; Li, Z.K. Whole Plant Responses, Key Processes, and Adaptation to Drought Stress: The Case of Rice. J. Exp. Bot. 2007, 58, 169–175.
- Zeid, I.M.; Shedeed, Z.A. Response of Alfalfa to Putrescine Treatment under Drought Stress. Biol. Plant. 2006, 50, 635–640.
- Shi, J.-F.; Mao, X.-G.; Jing, R.-L.; Pang, X.-B.; Wang, Y.-G.; Chang, X.-P. Gene Expression Profiles of Response to Water Stress at the Jointing Stage in Wheat. Agric. Sci. China 2010, 9, 325–330.
- Devnarain, N.; Crampton, B.G.; Chikwamba, R.; Becker, J.V.W.; O’Kennedy, M.M. Physiological Responses of Selected African Sorghum Landraces to Progressive Water Stress and Re-Watering. S. Afr. J. Bot. 2016, 103, 61–69.
- Tang, S.; Li, L.; Wang, Y.; Chen, Q.; Zhang, W.; Jia, G.; Zhi, H.; Zhao, B.; Diao, X. Genotype-Specific Physiological and Transcriptomic Responses to Drought Stress in Setaria Italica (an Emerging Model for Panicoideae Grasses). Sci. Rep. 2017, 7, 10009.
- Satish, L.; Rency, A.S.; Ramesh, M. Spermidine Sprays Alleviate the Water Deficit-Induced Oxidative Stress in Finger Millet (Eleusine Coracana L. Gaertn.) Plants. 3 Biotech 2018, 8, 63.
- Barzana, G.; Rios, J.J.; Lopez-Zaplana, A.; Nicolas-Espinosa, J.; Yepes-Molina, L.; Garcia-Ibañez, P.; Carvajal, M. Interrelations of Nutrient and Water Transporters in Plants under Abiotic Stress. Physiol. Plant. 2021, 171, 595–619.
- Carmody, M.; Waszczak, C.; Idänheimo, N.; Saarinen, T.; Kangasjärvi, J. ROS Signalling in a Destabilised World: A Molecular Understanding of Climate Change. J. Plant Physiol. 2016, 203, 69–83.
- Wang, J.; Li, C.; Li, L.; Reynolds, M.; Mao, X.; Jing, R. Exploitation of Drought Tolerance-Related Genes for Crop Improvement. Int. J. Mol. Sci. 2021, 22, 10265.
- Alhaithloul, H.A.S. Impact of Combined Heat and Drought Stress on the Potential Growth Responses of the Desert Grass Artemisia Sieberi Alba: Relation to Biochemical and Molecular Adaptation. Plants 2019, 8, 416.
- Recchia, G.H.; Caldas, D.G.G.; Beraldo, A.L.A.; Da Silva, M.J.; Tsai, S.M. Transcriptional Analysis of Drought-Induced Genes in the Roots of a Tolerant Genotype of the Common Bean (Phaseolus Vulgaris L.). Int. J. Mol. Sci. 2013, 14, 7155–7179.
- Hatfield, J.L.; Prueger, J.H. Temperature Extremes: Effect on Plant Growth and Development. Weather Clim. Extrem. 2015, 10, 4–10.
- Rai, A.; Rai, G.K.; Dubey, R.S. Heat Stress and Its Effects on Plant Growth and Metabolism. In Abiotic Stress Tolerance Mechanisms in Plants; CRC Press: Boca Raton, FL, USA, 2021; pp. 203–265. ISBN 1003163831.
- Jajoo, A.; Allakhverdiev, S.I. High-Temperature Stress in Plants: Consequences and Strategies for Protecting Photosynthetic Machinery. Plant Stress Physiol. 2017, 2017, 138–154.
- Chalanika De Silva, H.C.; Asaeda, T. Effects of Heat Stress on Growth, Photosynthetic Pigments, Oxidative Damage and Competitive Capacity of Three Submerged Macrophytes. J. Plant Interact. 2017, 12, 228–236.
- Yang, H.; Gu, X.; Ding, M.; Lu, W.; Lu, D. Heat Stress during Grain Filling Affects Activities of Enzymes Involved in Grain Protein and Starch Synthesis in Waxy Maize. Sci. Rep. 2018, 8, 15665.
- Bita, C.E.; Gerats, T. Plant Tolerance to High Temperature in a Changing Environment: Scientific Fundamentals and Production of Heat Stress-Tolerant Crops. Front. Plant Sci. 2013, 4, 273.
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80.
- Roy, S.J.; Negrão, S.; Tester, M. Salt Resistant Crop Plants. Curr. Opin. Biotechnol. 2014, 26, 115–124.
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of Salinity Stress on Plants and Its Tolerance Strategies: A Review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075.
- Muneer, S.; Park, Y.G.; Manivannan, A.; Soundararajan, P.; Jeong, B.R. Physiological and Proteomic Analysis in Chloroplasts of Solanum Lycopersicum L. under Silicon Efficiency and Salinity Stress. Int. J. Mol. Sci. 2014, 15, 21803–21824.
- Qados, A.M.S.A. Effect of Salt Stress on Plant Growth and Metabolism of Bean Plant Vicia Faba (L.). J. Saudi Soc. Agric. Sci. 2011, 10, 7–15.
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating Physiological Responses of Plants to Salinity Stress. Ann. Bot. 2017, 119, 1–11.
- Nxele, X.; Klein, A.; Ndimba, B.K. Drought and Salinity Stress Alters ROS Accumulation, Water Retention, and Osmolyte Content in Sorghum Plants. S. Afr. J. Bot. 2017, 108, 261–266.
- Jha, Y.; Subramanian, R.B. Regulation of Plant Physiology and Antioxidant Enzymes for Alleviating Salinity Stress by Potassium-Mobilizing Bacteria. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2016; pp. 149–162.
- Suriyan, C.-U.; Supaibulwattana, K.; Kirdmanee, C. Comparative Effects of Salt Stress and Extreme PH Stress Combined on Glycinebetaine Accumulation, Photosynthetic Abilities and Growth Characters of Two Rice Genotypes. Rice Sci. 2009, 16, 274–282.
- Sahi, C.; Singh, A.; Kumar, K.; Blumwald, E.; Grover, A. Salt Stress Response in Rice: Genetics, Molecular Biology, and Comparative Genomics. Funct. Integr. Genom. 2006, 6, 263–284.
- Swami, A.K.; Alam, S.I.; Sengupta, N.; Sarin, R. Differential Proteomic Analysis of Salt Stress Response in Sorghum Bicolor Leaves. Environ. Exp. Bot. 2011, 71, 321–328.
- Almodares, A.; Hadi, M.R.; Dosti, B. Effects of Salt Stress on Germination Percentage and Seedling Growth in Sweet Sorghum Cultivars. J. Biol. Sci. 2007, 7, 1492–1495.
- Gill, P.K.; Sharma, A.D.; Singh, P.; Bhullar, S.S. Changes in Germination, Growth and Soluble Sugar Contents of Sorghum Bicolor (L.) Moench Seeds under Various Abiotic Stresses. Plant Growth Regul. 2003, 40, 157–162.
- Sidhu, G.P.S.; Bali, A.S.; Singh, H.P.; Batish, D.R.; Kohli, R.K. Insights into the Tolerance and Phytoremediation Potential of Coronopus Didymus L.(Sm) Grown under Zinc Stress. Chemosphere 2020, 244, 125350.
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen Species, Abiotic Stress and Stress Combination. Plant J. 2017, 90, 856–867.
- Sandalio, L.M.; Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Luis, A. Role of Peroxisomes as a Source of Reactive Oxygen Species (ROS) Signaling Molecules. Peroxisomes Key Role Cell. Signal. Metab. 2013, 69, 231–255.
- Pei, Z.-M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium Channels Activated by Hydrogen Peroxide Mediate Abscisic Acid Signalling in Guard Cells. Nature 2000, 406, 731–734.
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930.
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Al Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681.
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800.
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, MA, USA, 2015; ISBN 0198717482.
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J. Lack of GLYCOLATE OXIDASE1, but Not GLYCOLATE OXIDASE2, Attenuates the Photorespiratory Phenotype of CATALASE2-Deficient Arabidopsis. Plant Physiol. 2016, 171, 1704–1719.
- Caverzan, A.; Casassola, A.; Brammer, S.P. Antioxidant Responses of Wheat Plants under Stress. Genet. Mol. Biol. 2016, 39, 1–6.
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Al Mahmud, J.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384.
- Gill, S.S.; Anjum, N.A.; Gill, R.; Yadav, S.; Hasanuzzaman, M.; Fujita, M.; Mishra, P.; Sabat, S.C.; Tuteja, N. Superoxide Dismutase—Mentor of Abiotic Stress Tolerance in Crop Plants. Environ. Sci. Pollut. Res. 2015, 22, 10375–10394.
- Pradedova, E.V.; Isheeva, O.D.; Salyaev, R.K. Superoxide Dismutase of Plant Cell Vacuoles. Biochem. Suppl. Ser. Membr. Cell Biol. 2009, 3, 24–32.
- Krönoiger, W.; Rennenberg, H.; Polle, A. Purification of Two Superoxide Dismutase Isozymes and Their Subcellular Localization in Needles and Roots of Norway Spruce (Picea Abies L.) Trees. Plant Physiol. 1992, 100, 334–340.
- Reddy, C.D.; Venkaiah, B. Isoenzymes of Superoxide Dismutase from Mung Bean (Phaseolus Aureus) Seedlings. Curr. Sci. 1982, 51, 987–988.
- Sharma, P.; Dubey, R.S. Drought Induces Oxidative Stress and Enhances the Activities of Antioxidant Enzymes in Growing Rice Seedlings. Plant Growth Regul. 2005, 46, 209–221.
- Khanna-Chopra, R.; Selote, D.S. Acclimation to Drought Stress Generates Oxidative Stress Tolerance in Drought-Resistant than-Susceptible Wheat Cultivar under Field Conditions. Environ. Exp. Bot. 2007, 60, 276–283.
- Zhang, J.; Kirkham, M.B. Antioxidant Responses to Drought in Sunflower and Sorghum Seedlings. New Phytol. 1996, 132, 361–373.
- Boeckx, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol Oxidase in Leaves: Is There Any Significance to the Chloroplastic Localization? J. Exp. Bot. 2015, 66, 3571–3579.
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic Stress Tolerance in Plants: Myriad Roles of Ascorbate Peroxidase. Front. Plant Sci. 2017, 8, 581.
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of Crop Wild Relatives: Expanding the Gene Pool for Crop Improvement. Plant Biotechnol. J. 2016, 14, 1070–1085.
- D’Agostino, N.; Tripodi, P. NGS-Based Genotyping, High-Throughput Phenotyping and Genome-Wide Association Studies Laid the Foundations for next-Generation Breeding in Horticultural Crops. Diversity 2017, 9, 38.
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.-S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X. A Draft Sequence of the Rice Genome (Oryza Sativa L. Ssp. Indica). Science 2002, 296, 79–92.
- Goff, S.A.; Ricke, D.; Lan, T.-H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H. A Draft Sequence of the Rice Genome (Oryza Sativa L. Ssp. Japonica). Science 2002, 296, 92–100.
- Project, I.R.G.S. The Map-Based Sequence of the Rice Genome. Nature 2005, 436, 793–800.
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115.
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A. The Sorghum Bicolor Genome and the Diversification of Grasses. Nature 2009, 457, 551–556.
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W. Genome Sequence of Foxtail Millet (Setaria Italica) Provides Insights into Grass Evolution and Biofuel Potential. Nat. Biotechnol. 2012, 30, 549–554.
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J. Reference Genome Sequence of the Model Plant Setaria. Nat. Biotechnol. 2012, 30, 555–561.
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D. Analysis of the Bread Wheat Genome Using Whole-Genome Shotgun Sequencing. Nature 2012, 491, 705–710.
- Mayer, K.F.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Muehlbauer, G.J. A Physical, Genetic and Functional Sequence Assembly of the Barley Genome. Int. Barley Genome Seq. Consort. 2012, 491, 711–716.
- Hatakeyama, M.; Aluri, S.; Balachadran, M.T.; Sivarajan, S.R.; Patrignani, A.; Grüter, S.; Poveda, L.; Shimizu-Inatsugi, R.; Baeten, J.; Francoijs, K.-J. Multiple Hybrid de Novo Genome Assembly of Finger Millet, an Orphan Allotetraploid Crop. DNA Res. 2018, 25, 39–47.
- Braslavsky, I.; Hebert, B.; Kartalov, E.; Quake, S.R. Sequence Information Can Be Obtained from Single DNA Molecules. Proc. Natl. Acad. Sci. USA 2003, 100, 3960–3964.
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323, 133–138.
- Levene, M.J.; Korlach, J.; Turner, S.W.; Foquet, M.; Craighead, H.G.; Webb, W.W. Zero-Mode Waveguides for Single-Molecule Analysis at High Concentrations. Science 2003, 299, 682–686.
- Greenleaf, W.J.; Block, S.M. Single-Molecule, Motion-Based DNA Sequencing Using RNA Polymerase. Science 2006, 313, 801.
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F. Systems Biology Approach in Plant Abiotic Stresses. Plant Physiol. Biochem. 2017, 121, 58–73.
- Zhu, L.; Wu, H.; Li, H.; Tang, H.; Zhang, L.; Xu, H.; Jiao, F.; Wang, N.; Yang, L. Short Tandem Repeats in Plants: Genomic Distribution and Function Prediction. Electron. J. Biotechnol. 2021, 50, 37–44.
- Feuillet, C.; Stein, N.; Rossini, L.; Praud, S.; Mayer, K.; Schulman, A.; Eversole, K.; Appels, R. Integrating Cereal Genomics to Support Innovation in the Triticeae. Funct. Integr. Genom. 2012, 12, 573–583.
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186.
- Mao, L.; Chen, M.; Chu, Q.; Jia, L.; Sultana, M.H.; Wu, D.; Kong, X.; Qiu, J.; Ye, C.-Y.; Zhu, Q.-H. RiceRelativesGD: A Genomic Database of Rice Relatives for Rice Research. Database 2019, 2019, baz110.
- McLaren, C.G.; Bruskiewich, R.M.; Portugal, A.M.; Cosico, A.B. The International Rice Information System. A Platform for Meta-Analysis of Rice Crop Data. Plant Physiol. 2005, 139, 637–642.
- Li, Y.; Xiao, J.; Chen, L.; Huang, X.; Cheng, Z.; Han, B.; Zhang, Q.; Wu, C. Rice Functional Genomics Research: Past Decade and Future. Mol. Plant 2018, 11, 359–380.
- Woodhouse, M.R.; Cannon, E.K.; Portwood, J.L.; Harper, L.C.; Gardiner, J.M.; Schaeffer, M.L.; Andorf, C.M. A Pan-Genomic Approach to Genome Databases Using Maize as a Model System. BMC Plant Biol. 2021, 21, 385.
- Lawrence, C.J.; Dong, Q.; Polacco, M.L.; Seigfried, T.E.; Brendel, V. MaizeGDB, the Community Database for Maize Genetics and Genomics. Nucleic Acids Res. 2004, 32, D393–D397.
- Cho, K.T.; Portwood, J.L.; Gardiner, J.M.; Harper, L.C.; Lawrence-Dill, C.J.; Friedberg, I.; Andorf, C.M. MaizeDIG: Maize Database of Images and Genomes. Front. Plant Sci. 2019, 10, 1050.
- Lawrence, C.J.; Harper, L.C.; Schaeffer, M.L.; Sen, T.Z.; Seigfried, T.E.; Campbell, D.A. MaizeGDB: The Maize Model Organism Database for Basic, Translational, and Applied Research. Int. J. Plant Genom. 2008, 2008, 496957.
- Shamimuzzaman, M.; Gardiner, J.M.; Walsh, A.T.; Triant, D.A.; Le Tourneau, J.J.; Tayal, A.; Unni, D.R.; Nguyen, H.N.; Portwood, J.L.; Cannon, E.K.S. MaizeMine: A Data Mining Warehouse for the Maize Genetics and Genomics Database. Front. Plant Sci. 2020, 11, 592730.
- Zhang, X.; Li, J.; Liu, A.; Zou, J.; Zhou, X.; Xiang, J.; Rerksiri, W.; Peng, Y.; Xiong, X.; Chen, X. Expression Profile in Rice Panicle: Insights into Heat Response Mechanism at Reproductive Stage. PLoS ONE 2012, 7, e49652.
- Hittalmani, S.; Mahesh, H.B.; Shirke, M.D.; Biradar, H.; Uday, G.; Aruna, Y.R.; Lohithaswa, H.C.; Mohanrao, A. Genome and Transcriptome Sequence of Finger Millet (Eleusine Coracana (L.) Gaertn.) Provides Insights into Drought Tolerance and Nutraceutical Properties. BMC Genom. 2017, 18, 465.
- Xiao, C.-L.; Chen, Y.; Xie, S.-Q.; Chen, K.-N.; Wang, Y.; Han, Y.; Luo, F.; Xie, Z. MECAT: Fast Mapping, Error Correction, and de Novo Assembly for Single-Molecule Sequencing Reads. Nat. Methods 2017, 14, 1072–1074.
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W. The Genome of Broomcorn Millet. Nat. Commun. 2019, 10, 436.
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L. Echinochloa Crus-Galli Genome Analysis Provides Insight into Its Adaptation and Invasiveness as a Weed. Nat. Commun. 2017, 8, 1031.
- Varshney, R.K.; Shi, C.; Thudi, M.; Mariac, C.; Wallace, J.; Qi, P.; Zhang, H.; Zhao, Y.; Wang, X.; Rathore, A. Pearl Millet Genome Sequence Provides a Resource to Improve Agronomic Traits in Arid Environments. Nat. Biotechnol. 2017, 35, 969–976.
- Sutton, G.G.; White, O.; Adams, M.D.; Kerlavage, A.R. TIGR Assembler: A New Tool for Assembling Large Shotgun Sequencing Projects. Genome Sci. Technol. 1995, 1, 9–19.
- Myers, E.W. The Fragment Assembly String Graph. Bioinformatics 2005, 21, ii79–ii85.
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z. Genome Sequencing in Microfabricated High-Density Picolitre Reactors. Nature 2005, 437, 376–380.
- Warren, R.L.; Sutton, G.G.; Jones, S.J.M.; Holt, R.A. Assembling Millions of Short DNA Sequences Using SSAKE. Bioinformatics 2007, 23, 500–501.
- Jeck, W.R.; Reinhardt, J.A.; Baltrus, D.A.; Hickenbotham, M.T.; Magrini, V.; Mardis, E.R.; Dangl, J.L.; Jones, C.D. Extending Assembly of Short DNA Sequences to Handle Error. Bioinformatics 2007, 23, 2942–2944.
- Dohm, J.C.; Lottaz, C.; Borodina, T.; Himmelbauer, H. SHARCGS, a Fast and Highly Accurate Short-Read Assembly Algorithm for de Novo Genomic Sequencing. Genome Res. 2007, 17, 1697–1706.
- Butler, J.; MacCallum, I.; Kleber, M.; Shlyakhter, I.A.; Belmonte, M.K.; Lander, E.S.; Nusbaum, C.; Jaffe, D.B. ALLPATHS: De Novo Assembly of Whole-Genome Shotgun Microreads. Genome Res. 2008, 18, 810–820.
- Hernandez, D.; François, P.; Farinelli, L.; Østerås, M.; Schrenzel, J. De Novo Bacterial Genome Sequencing: Millions of Very Short Reads Assembled on a Desktop Computer. Genome Res. 2008, 18, 802–809.
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829.
- Miller, J.R.; Delcher, A.L.; Koren, S.; Venter, E.; Walenz, B.P.; Brownley, A.; Johnson, J.; Li, K.; Mobarry, C.; Sutton, G. Aggressive Assembly of Pyrosequencing Reads with Mates. Bioinformatics 2008, 24, 2818–2824.
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123.
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303.
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y. Erratum: SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. Gigascience 2015, 4, 1.
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877.
- Duan, C.; Argout, X.; Gébelin, V.; Summo, M.; Dufayard, J.-F.; Leclercq, J.; Piyatrakul, P.; Pirrello, J.; Rio, M.; Champion, A. Identification of the Hevea BrasiliensisAP2/ERF Superfamily by RNA Sequencing. BMC Genom. 2013, 14, 30.
- Yang, Y.; Smith, S.A. Optimizing de Novo Assembly of Short-Read RNA-Seq Data for Phylogenomics. BMC Genom. 2013, 14, 328.
- Wang, W.; Ding, G.-D.; White, P.J.; Wang, X.-H.; Jin, K.-M.; Xu, F.-S.; Shi, L. Mapping and Cloning of Quantitative Trait Loci for Phosphorus Efficiency in Crops: Opportunities and Challenges. Plant Soil 2019, 439, 91–112.
- Rashid, B.; Husnain, T.; Riazuddin, S. Genomic Approaches and Abiotic Stress Tolerance in Plants. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1–37.
- Priyadarshan, P.M. Breeding for Abiotic Stress Adaptation. In Plant Breeding: Classical to Modern; Springer: Berlin/Heidelberg, Germany, 2019; pp. 413–455.
- Kumar, S.; Bhati, J.; Saha, A.; Lal, S.B.; Pandey, P.K.; Mishra, D.C.; Farooqi, M.S.; Kumar, A.; Chaturvedi, K.K.; Rai, A. CerealESTDb: A Comprehensive Resource for Abiotic Stress-Responsive Annotated ESTs with Predicted Genes, Gene Ontology, and Metabolic Pathways in Major Cereal Crops. Front. Genet. 2022, 13, 842868.
- Riaz, A.; Kanwal, F.; Börner, A.; Pillen, K.; Dai, F.; Alqudah, A.M. Advances in Genomics-Based Breeding of Barley: Molecular Tools and Genomic Databases. Agronomy 2021, 11, 894.
- Udoh, L.I.; Obaseojei, W.P.; Uzoebo, C. Single Nucleotide Polymorphisms: A Modern Tool to Screen Plants for Desirable Traits. In Plant Breeding-Current and Future Views; IntechOpen: London, UK, 2021; ISBN 1839683104.
- Bhattarai, G.; Shi, A.; Kandel, D.R.; Solís-Gracia, N.; da Silva, J.A.; Avila, C.A. Genome-Wide Simple Sequence Repeats (SSR) Markers Discovered from Whole-Genome Sequence Comparisons of Multiple Spinach Accessions. Sci. Rep. 2021, 11, 9999.
- Yonemaru, J.; Yamamoto, T.; Fukuoka, S.; Uga, Y.; Hori, K.; Yano, M. Q-TARO: QTL Annotation Rice Online Database. Rice 2010, 3, 194–203.
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y. Development and Mapping of 2240 New SSR Markers for Rice (Oryza Sativa L.). DNA Res. 2002, 9, 199–207.
- Ganie, S.A.; Borgohain, M.J.; Kritika, K.; Talukdar, A.; Pani, D.R.; Mondal, T.K. Assessment of Genetic Diversity of Saltol QTL among the Rice (Oryza Sativa L.) Genotypes. Physiol. Mol. Biol. Plants 2016, 22, 107–114.
- Ghimire, K.H.; Quiatchon, L.A.; Vikram, P.; Swamy, B.P.M.; Dixit, S.; Ahmed, H.; Hernandez, J.E.; Borromeo, T.H.; Kumar, A. Identification and Mapping of a QTL (QDTY1. 1) with a Consistent Effect on Grain Yield under Drought. Field Crops Res. 2012, 131, 88–96.
- Tang, W.; Wu, T.; Ye, J.; Sun, J.; Jiang, Y.; Yu, J.; Tang, J.; Chen, G.; Wang, C.; Wan, J. SNP-Based Analysis of Genetic Diversity Reveals Important Alleles Associated with Seed Size in Rice. BMC Plant Biol. 2016, 16, 93.
- Rauf, S.; Al-Khayri, J.M.; Zaharieva, M.; Monneveux, P.; Khalil, F. Breeding Strategies to Enhance Drought Tolerance in Crops. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Springer: Berlin/Heidelberg, Germany, 2016; pp. 397–445.
- Wilkinson, P.A.; Winfield, M.O.; Barker, G.L.A.; Allen, A.M.; Burridge, A.; Coghill, J.A.; Edwards, K.J. CerealsDB 2.0: An Integrated Resource for Plant Breeders and Scientists. BMC Bioinform. 2012, 13, 219.
- Kayıhan, C.; Eyidoğan, F. Omics in Oxidative Stress Tolerance in Crops. React. Oxyg. Nitrogen Sulfur Species Plants Prod. Metab. Signal. Def. Mech. 2019, 195–224.
- McNally, K.L.; Childs, K.L.; Bohnert, R.; Davidson, R.M.; Zhao, K.; Ulat, V.J.; Zeller, G.; Clark, R.M.; Hoen, D.R.; Bureau, T.E. Genomewide SNP Variation Reveals Relationships among Landraces and Modern Varieties of Rice. Proc. Natl. Acad. Sci. USA 2009, 106, 12273–12278.
- Chen, J.B.; Jing, R.L.; Yuan, H.Y.; Wei, B.; Chang, X.P. Single Nucleotide Polymorphism of TaDREB1 Gene in Wheat Germplasm. Sci. Agric. Sin. 2005, 38, 2387–2394.
- Garg, D.; Sareen, S.; Dalal, S.; Tiwari, R.; Singh, R. Heat Shock Protein Based SNP Marker for Terminal Heat Stress in Wheat (Triticum Aestivum L.). Aust. J. Crop Sci. 2012, 6, 1516–1521.
- Setter, T.L.; Yan, J.; Warburton, M.; Ribaut, J.-M.; Xu, Y.; Sawkins, M.; Buckler, E.S.; Zhang, Z.; Gore, M.A. Genetic Association Mapping Identifies Single Nucleotide Polymorphisms in Genes That Affect Abscisic Acid Levels in Maize Floral Tissues during Drought. J. Exp. Bot. 2011, 62, 701–716.
- Assenov, B.; Andjelkovic, V.; Ignjatovic-Micic, D.; Vancetovic, J.; Nikolic, A.; Christov, N.K.; Tsonev, S.; Abu-Mhadi, N.; Vassilev, D.; Muhovski, Y. Identification of SNP Mutations in MYBE-1 Gene Involved in Drought Stress Tolerance in Maize. Bulg. J. Agric. Sci 2013, 19, 181–185.
- Close, T.J.; Wanamaker, S.I.; Caldo, R.A.; Turner, S.M.; Ashlock, D.A.; Dickerson, J.A.; Wing, R.A.; Muehlbauer, G.J.; Kleinhofs, A.; Wise, R.P. A New Resource for Cereal Genomics: 22K Barley GeneChip Comes of Age. Plant Physiol. 2004, 134, 960–968.
- Stein, N.; Prasad, M.; Scholz, U.; Thiel, T.; Zhang, H.; Wolf, M.; Kota, R.; Varshney, R.K.; Perovic, D.; Grosse, I. A 1000-Loci Transcript Map of the Barley Genome: New Anchoring Points for Integrative Grass Genomics. Theor. Appl. Genet. 2007, 114, 823–839.
- Sato, K.; Nankaku, N.; Takeda, K. A High-Density Transcript Linkage Map of Barley Derived from a Single Population. Heredity 2009, 103, 110–117.
- Elango, D.; Sandoya, G.; Chopra, S. Techniques and Tools of Modern Plant Breeding. In Plant Biotechnology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 17–26.
- Chang, J.; Tian, J.; Yang, Y.; Zhong, R.; Li, J.; Zhai, K.; Ke, J.; Lou, J.; Chen, W.; Zhu, B. A Rare Missense Variant in TCF7L2 Associates with Colorectal Cancer Risk by Interacting with a GWAS-Identified Regulatory Variant in the MYC EnhancerAn Exome-Wide Association Study of Colorectal Cancer. Cancer Res. 2018, 78, 5164–5172.
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A Tutorial on Conducting Genome-wide Association Studies: Quality Control and Statistical Analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608.
- Huang, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; Li, M. Genome-Wide Association Studies of 14 Agronomic Traits in Rice Landraces. Nat. Genet. 2010, 42, 961–967.
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E. Population Genomic and Genome-Wide Association Studies of Agroclimatic Traits in Sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458.
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H. A Haplotype Map of Genomic Variations and Genome-Wide Association Studies of Agronomic Traits in Foxtail Millet (Setaria Italica). Nat. Genet. 2013, 45, 957–961.
More
