Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Asunción Contreras and Version 2 by Camila Xu.
The pyridoxal phosphate-binding protein (PLPBP) family (also termed ProsC/PROSC or COG0325 family) members are found in all kingdoms of life, exemplified by the proteins YBL036C (yeast), YggS (Gram-negative bacteria), YlmE (Gram-positive bacteria), PipY (cyanobacteria), PLPHP (humans) and HTH5 (rice).
- cyanobacteria
- nitrogen regulation
- COG0325
- PLPHP
- PLPBP
1. Introduction
Cyanobacteria, phototrophic organisms performing oxygenic photosynthesis, constitute an ecologically and biotechnologically important phylum, responsible for the evolution of the oxygenic atmosphere, being the main contributors to marine primary production [1]. Their photosynthetic lifestyle and ease of cultivation make them ideal production systems for several high-value compounds, including biofuels [2]. Despite important breakthroughs in the genetic analysis of cyanobacteria, there is still a remarkable proportion of genes of unknown function in this phylum, many of which are presumably relevant to the biology of cyanobacteria.
The cyanobacterium Synechococcus elongatus PCC7942 (hereafter S. elongatus), the only photosynthetic organism for which the contribution of each gene to fitness has been evaluated so far [3], is being used as a model system to address fundamental questions concerning the photosynthetic lifestyle. More recently, the S. elongatus genome has been used as the reference organism to create a database for the Cyanobacterial-Linked Genome [4], accessible through an interactive platform “https://dfgm.ua.es/es/cyanobacterial-genetics/dclg/index.htm (accesed on 1 August 2022)”.
In bacteria and plants, 2-oxoglutarate (2-OG), a key metabolic signal of the intracellular carbon-to-nitrogen balance, is sensed by the highly conserved and widely distributed signal transduction protein PII. PII regulates the activity of proteins involved in nitrogen metabolism by direct protein–protein interactions [5]. In S. elongatus PII interacts with a small (89 residues) protein called PipX (PII-interacting protein X), which was initially identified in yeast two-hybrid analyses [6,7].
PipX was also found in searches for proteins interacting with NtcA, the global transcriptional regulator involved in nitrogen assimilation in cyanobacteria [8]. PipX stabilizes the conformation of NtcA which is transcriptionally active and probably helps the local recruitment of RNA polymerase to NtcA-dependent promoters [9]. At low 2-OG concentrations corresponding to nitrogen-excess conditions, the sequestration of PipX by PII renders PipX unavailable for NtcA binding and activation, reducing the expression of NtcA-dependent gene targets [9,10,11,12,13]. Partner swapping by PipX is enabled by its N-terminal Tudor-like domain (TLD/KOW), which provides contacts for both NtcA and PII. Complex formation with PipX increases the affinity of PII for ADP [9], and, conversely, the interaction between PII and PipX is highly sensitive to fluctuations in the ATP/ADP ratio [14]. Thus, PipX partner swapping between PII and NtcA integrates signaling of the carbon-to-nitrogen ratio and the energy status by PII with the regulation of nitrogen-responsive genes controlled by NtcA [10,15,16].
Interestingly, a high PipX/PII ratio prevents growth [11,17] and, consistent with this, cyanobacterial genomes always contain at least as many copies of glnB as of pipX [18], suggesting that a relatively high ratio of PII over PipX is required to counteract unwanted interactions with low-affinity PipX partners.
In S. elongatus pipX is co-transcribed with the downstream gene pipY. This last gene belongs to the widely distributed and highly conserved pyridoxal phosphate (PLP)-binding protein (COG0325/PLPBP) family that is involved in vitamin B6 and amino acid homeostasis [19]. The PLPBP family (also termed ProsC/PROSC or COG0325 family) members are found in all kingdoms of life, exemplified by the proteins YBL036C (yeast), YggS (Gram-negative bacteria), YlmE (Gram-positive bacteria), PipY (cyanobacteria), PLPHP (humans) and HTH5 (rice). These are all single-domain proteins exhibiting the fold type III of PLP-holoenzymes [20,21,22,23,24] with no known enzymatic activity.
2. Structural and Functional Features of PLPBPs
2.1. PLP Is Solvent-Exposed in PLPBP Structures
The vitamin B6 vitamer PLP is used as a cofactor for enzyme-catalyzed reactions which include transamination, decarboxylation, racemization, aldol cleavage, or replacement reactions among others [26]. Since amino acid metabolism and other essential processes require PLP-dependent enzymes [27,28], PLP availability is of paramount importance to supply cofactors to activate newly synthesized apo-B6 enzymes. PLP is also required as a cofactor of glycogen phosphorylase [29] and certain transcriptional factors and regulators [28]. However, its aldehyde group endows PLP with high chemical reactivity, sometimes causing the inactivation of proteins (see for example, [30]), and therefore additional mechanisms are required for keeping the levels of free PLP low in cells and tissues. In the first report of a member of this family, Eswaramoorthy et al. (2003) documented structural parallelisms between the yeast protein YBL036C and the N-terminal domain of alanine racemases, leading them to infer (and even to provide some experimental hints for it) that PLPBP had alanine racemase activity [21]. However, no amino acid racemase, decarboxylase, deaminase, or transaminase activities were found for E. coli or human proteins [31], and although crystal structures of alanine racemase with bound substrates (D-ala) or inhibitors (D-cycloserine) have been determined [32], extensive crystallization attempts with these molecules did not detect any binding to PipY [22]. Furthermore, in vivo work did not support alanine racemase activity for S. elongatus PipY [19]. Therefore, despite the key importance of the PLP cofactor for PLPBP function (see below), PLP appears to have no catalytic function in the PLPBP family. Structures of six PLPBP members have been determined and deposited in the Protein DataBank (PDB, “https://www.rcsb.org/ (accesed on 1 September 2022)”) (Table 1). All of these structures correspond to single-domain chains folded according to the triose phosphate isomerase (TIM) barrel typically found in the fold type III of PLP-dependent enzymes. The only ones reported to date from a eukaryotic organism correspond to yeast protein YBL036C. The others are from a Gram-positive bacterium (Bifidobacterium adolescentis), and four Gram-negative bacteria including the cyanobacterium S. elongatus (Table 1). S. elongatus PipY structures with and without PLP offer high resolution and have been used to estimate the effects of clinical missense mutations found in the PLPBP human gene in patients with vitamin B6-dependent epilepsy [22,23]. Here, we use PipY as a reference for the additional discussion on structural and functional details concerning studied members of the protein family. Figure 1 shows the structure of PipY containing PLP (PDB file 5NM8).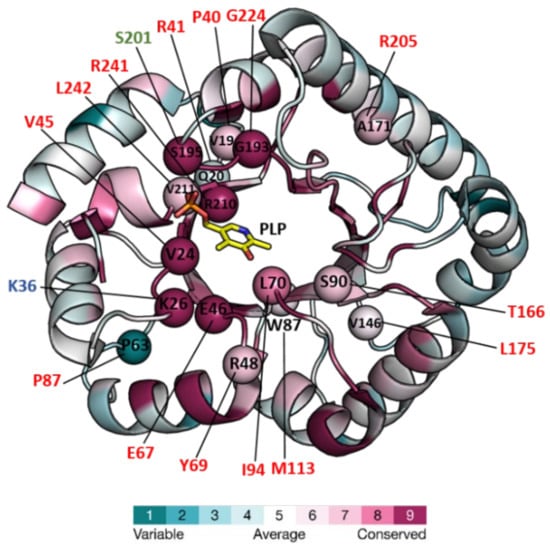
Figure 1. Structure of PipY from S. elongatus (PDB 5NM8) colored for the evolutionary conservation of residues among PLPBP homologs, and mapping therein, residues targeted by missense mutations. The structure is in a cartoon representation except for the PLP, which is in a stick representation with C, O, N, and P atoms in yellow, red, blue, and orange, respectively. Color-coding of the structure from cyan to magenta according to the residue conservation score (the higher, the more conserved) given by The ConSurf Server “URL https://consurf.tau.ac.il/consurf_index.php (accesed on 3 August 2022) when queried with chain A of the PDB 5NM8, with default parameters. Spheres mark the location in PipY of known human PLPHP mutations (see Table 2). Residue numbers are given in one letter code, in black for S. elongatus, and shown in red, green, and blue, the human mutations causing vitamin B6-dependent epilepsy, and the in vitro mutations obtained in the corresponding proteins of F. nucleatum, and E. coli, respectively.
Table 1. Structures of COG0325/PLPB family proteins were determined and deposited in the Protein DataBank (PDB).
| Organism | Protein | PDB File | Vitamer | Ligands | Amino Acid Changes | Resolut. (Å) |
Deposition Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | YggS | 1W8G | PLP | Isocitrate | None | 2.00 | 2004 | – |
| 3SY1 | PLP | MES Acetate |
L32V/G56S/N58H/H81N/ I83A/H102I/M165S/S202A/ M205Q/R221A hexamutant |
1.47 | 2011 | – | ||
| 7UBQ | PNP * | None | None | 2.60 | 2022 | – | ||
| 7UB4 | PLP | None | K36A/K38A/K233A/K234 | 2.40 | 2022 | – | ||
| 7UAX | None | PO4H3 | K36A/K38A | 2.07 | 2022 | |||
| None | ||||||||
| 2.10 | ||||||||
| 1999 | ||||||||
| [ | ||||||||
| 21 | ||||||||
| ] | ||||||||
* Pyridoxine-5′-phosphate. ** Methionine replaced by selenomethionine—data not available.
The TIM-barrel fold, initially described for triosephosphate isomerase [33], is a highly widespread protein fold, generally reported as consisting of eight α helices that alternate with parallel β strands of a circularly closed β-sheet, in which the helices encircle the sheet (reviewed in [34]). The TIM-barrel of PLP proteins, first described for alanine racemase [32], characterizes the fold type III of PLP-dependent enzymes and presents an extra N-terminal α helix preceding the first of the eight βα repeats. However, while this modified TIM-barrel is part of a two-domain subunit forming homodimers in alanine racemase, ornithine decarboxylase, and the broad specificity amino acid racemase [20,26,35], PLPBP members are single-domain proteins that appear to be mainly monomers (but see discussion below) ([21,22]; and other PDBs in Table 1).
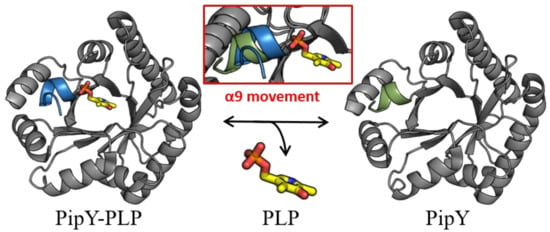
While PLP is found in PLPBP structures in the same location that it is found in the fold type III of PLP enzymes, in all PLPBP structures the PLP cofactor is solvent-exposed and highly accessible, thus being appropriately positioned for a role of PLPBP as a PLP delivering device in cells. In this context, the role of the C-terminal α helix (helix 9) of PipY in anchoring the phosphate of PLP, and the relatively large changes in helix 9 orientation depending on the presence or absence of PLP have led to the hypothesizing [22] that this helix may have a role in being a trigger for the binding and release of PLP (Figure 2).
 The available data so far suggest that the proteins of this family might act as PLP carriers which supply the cofactor to PLP-dependent enzymes, shielding this cofactor from unwanted reactions with other molecules, although this has not been strictly proven and the mechanisms involved remain unclarified.
The available data so far suggest that the proteins of this family might act as PLP carriers which supply the cofactor to PLP-dependent enzymes, shielding this cofactor from unwanted reactions with other molecules, although this has not been strictly proven and the mechanisms involved remain unclarified.

Figure 2. PipY structures with and without PLP illustrate the two positions of helix 9. Cartoon representation of PipY structure from S. elongatus complexed with PLP (PDB 5NM8) and PipY-Apo form (PBD 5NLC), with α-helix 9 shown in blue and green, respectively. Inset: Displacement of α-helix 9 observed in the PLP-containing form is highlighted by superimposing both protein forms. The PLP molecule is illustrated using stick representation, where C, O, N, and P atoms are colored yellow, red, blue, and orange, respectively.
2.2. Dimerization of Just Some PLPBP Family Members?
While all available crystal structures are consistent with PLPBP family members being monomers, size-exclusion chromatography of human PLPHP, performed in two different studies [23,36] revealed a second peak corresponding to dimers. In [23], the minor peak was shown to depend on disulfide bridges, a result interpreted as PLPHP being mainly monomeric with the possibility of stable dimerization via the formation of a disulfide bridge between exposed cysteines. Consistent with this, human mutation Tyr69Cys increased dimer formation (Table 2). However, Fux and Sieber [36] challenged this view, reporting that their PLPHP preparation was predominantly dimeric even under reducing conditions [36]. They also suggested that discrepancies with the previous work may be due to differences in expression strains or purification strategies. It is worth noting that while the human protein contains five cysteine residues, the amino acid chains of S. elongatus PipY and E. coli YggS have just one or two cysteines, respectively, and thus they would be less prone to making disulfide bonds under oxidative conditions. Table 2. Missense mutations reported in human PLPHP associated with vitamin B6-dependent epilepsy, as well as two experimental mutations in orthologous bacterial proteins. Molecular mechanisms of damage.| Amino Acid Change in HuPLPHP | Molecular Mechanism: Effect on PLPBP Protein | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid | Conserv. Score 1 | Change | Clinical effects | Reported in | Number of patients | Observed effect | Inferred from | Ref. | |||||||||
| P40 | 7 | P40L | Seizures | [44] [51] |
1 (P40L/R241Q) 1 (P40L/splicing) |
↓ thermostability | In vitro studies on rHuPLPHP | [23] | |||||||||
| R41 | 4 | R41Q | Seizures Mild disease 2 |
[54] [43] |
2 (homozygous;R41Q/V45D) 3 (homozygous) |
↓yield/misfolding? ↓thermostability |
In vitro studies on rHuPLPHP | [36] | |||||||||
| – | |||||||||||||||||
| R41W | Seizures, death | [50] | 1 (homozygous) | NT | NT | NT | |||||||||||
| V45 | 9 | V45D | Seizures | [54] | 1 (R41Q/V45D) | ↓↓PLP content ↓ thermostability |
In vitro studies on rHuPLPHP | [36] | 7U9H | None | SO4H | ||||||
| K47 | 2 | None | 2.00 | 2022 | 9 | – | |||||||||||
| K47A | Not reported in humans (prenatally lethal?) | lack of PLP | rEcyggS | K36A | [ | 31] | 7UBP | PLP | SO4H2 | K36A/K137A | 2.30 | ||||||
| E67 | 9 | E67K | Seizures Severe disease 2 |
[54 | 2022 | ] [43] |
1 (homozygous) | – | |||||||||
| 3 (homozygous) | Misfolding | In vitro studies on rHuPLPHP | [ | 36 | ] | 7UB8 | PLP | Butanediol | K38A/K137A/K233A/K234A | 2.30 | |||||||
| Y69 | 7 | Y69C | Seizures Moderate disease 2 |
[44] | 1 (homozygous) | Higher dimerization ↓PLP content | 2022 | – | |||||||||
| In vitro studies on rHuPLPHP | [ | 23 | ] | 7UAU | PLP | SO4H2 | K137A | 2.10 | 2022 | ||||||||
| P87 | – | ||||||||||||||||
| 1 | P87L | Seizures | Severe disease 2 |
[41] [44] [53] |
1(P87L/R241Q) 1 (homozygous) 1 (P87L/splicing) |
↓solubility/misfolding | In vitro studies on rHuPLPHP | [23] | 7UAT | PLP | PO4H3 | ||||||
| I94 | 8 | I94F | Seizures Mild disease 2 | K36A | [43 | 2.00 | ] | 1 (homozygous)2022 | – | ||||||||
| Proposed ↓ in PLP saturation | Structural modeling of HuPLPHP | [ | 43 | ] | 7U9C | PLP | |||||||||||
| M113 | 6 | M113T | PO | 4H3 | None | 2.10 | 2022 | – | |||||||||
| Seizures | [ | 51 | ] | 1 (M113T/C15X) | NT | NT | NT | Bifidobacterium adolescentis |
YggS | 3CPG | PLP | Acetate | Se-Met ** | 1.71 | 2008 | – | |
| T116 | 7 | T116I | Seizures Severe disease 2 |
[43] | 2 (1 homozygous; 1 homozygous (T116I/H275D)) |
Proposed ↓ in PLP saturation | Structural modeling of HuPLPHP | [43] | Agrobacterium tumefaciens | YggS | 3R79 | PLP | Acetate Pr+3 |
Se-Met ** | 1.90 | 2011 | – |
| L175 | 6 | L175P | Seizures Severe disease 2 |
[41] | 1 (homozygous) | Misfolding | In vitro studies on rHuPLPHP | [23] | Synechococcus elongatus | PipY | 5NLC | None | PO4H3 | None | 1.90 | 2017 | |
| R205 | 7 | R205Q | Seizures Moderate disease 2 |
[44] [54] |
1 (R205Q/null) 1 (homozygous) | [ | 22 | ] | |||||||||
| ↓thermostability | In vitro studies on rHuPLPHP | [ | 23 | ] | 5NM8 | PLP | Ca2+ | None | 1.93 | 2017 | [22] | ||||||
| G224 | 9 | G224A | Seizures Severe disease 2 |
[43] | 1 (G224A/splicing) | Proposed ↓ in PLP saturation | Structural modeling of HuPLPHP | [43] | Fusobacterium nucleatum | YggS | 7F8E | None | SO4H2 | Se-Met ** | 2.08 | 2021 | – |
| 6KZW | None | PO4H3 | T5A/N202S, Se-Met ** | 2.08 | 2019 | – | |||||||||||
| 7YGF | Structure not released | 2.08 | 2022 | [24] | |||||||||||||
| Saccharomyces cerevisiae | YBL036C | ||||||||||||||||
| S226 | 9 | S226A | Not reported in humans (prenatally lethal?) | ↓PLP saturation | rFnS201A | [24] | |||||||||||
| R241 | 9 | R241Q | Seizures | [41] [44] [52] |
1 (P87L/R241Q) 1 (P40L/R241Q) 1(R241Q/splicing) |
↓solubility ↓thermostability ↓PLP binding |
In vitro studies on rHuPLPHP In vitro studies on rSePipYR210Q |
[22,23] | |||||||||
| I242 | 7 | I242T | Seizures | [45] | 1 (homozygous) | NT | NT | NT | 1CT5 | PLP | None | ||||||
| H275 | NA | H275D | Se-Met ** | 2.00 | 1999 | [ | Seizures | 21] | |||||||||
| [ | 43 | ] | 1 (homozygous T116I/H275D) | Variant of uncertain significance (VUS) | Structural modeling of HuPLPHP | [ | 43 | ] | 1B54 | PLP | None | ||||||
NA: not applicable. NT: not tested. HuPLPHP: Human PLPHP. rHuPLPHP: recombinant human PLPHP. rEcyggSK36A: recombinant E. coli yggS K36A mutant. rSePipYR210Q: recombinant S. elongatus PipY R210Q mutant. rFnS201A: recombinant F. nucleatum yggS S201A mutant. 1 Conservation score as given in Figure 1. 2 Severity score given in [43].
Although the relative importance of dimeric forms of PLPBP family members in cell systems remains to be determined, it is worth noting that dimer formation could significantly enhance the putative function of PLPBPs in shielding PLP transport or storage, which are the most likely functions attributed to PLPBPs so far. Importantly, the identification of dimeric forms of human PLPHP opens the possibility that at least two different forms of PLPBPs (monomeric and dimeric), perhaps with different regulatory properties, might be found in cells. Whether dimer formation is a property of just some PLPBP members or a universal feature of the family, and whether in vivo dimer formation requires cysteines in critical positions or can also be induced by other effectors or cell components are open questions that require further investigation.
