Autism spectrum disorder (ASD) is a serious neurodevelopmental disorder characterized by the impairment of the cognitive function of a child. Studies suggested that the intestinal microbiota has a critical role in the function and regulation of the central nervous system, neuroimmune system, and neuroendocrine system. Any adverse changes in the gut-brain axis may cause serious diseases. Food preferences and dietary patterns are considered as key influencing factors of ASD development. Several recent reviews narrated the importance of dietary composition on controlling or reducing the ASD symptoms. It has been known that consumption of probiotics confers several health benefits by positive amendment of gut microbiota. Influence of probiotic intervention in children with ASD have also been reported and it has been considered as an alternative and complementary therapeutic supplement for ASD. The present manuscript discussed the role of microbiota in the development of ASD.
- Autism spectrum disorder
- Microbiome
- Cognition.
1. Introduction
Autism spectrum disorder (ASD) refers to a group of neurodevelopmental disorders characterized by the impairment of social interaction and communication skills with rigid and repetitive behaviors. Autism affects both children and adults, based on severity and intellectual ability; they may either lead a normal life or suffer a devastating disability requiring institutional care.[1] Distinctive symptoms of ASD are deficits in social behaviors and nonverbal interactions such as the avoidance of eye contact, inability to control emotion or understand the emotions of others, facial expression and body gestures in the first three years of life. Paul Eugen Bleuler is a swiss psychiatrist that coined the term “autism” (Greek word “autos” means “self”) for group of symptoms related to schizophrenia, while Hans Asperger and Leo Kanner designed the modern study of autism. An epidemiological survey revealed that ASD is the most prevalent non-immune mediated CNS disorder with an incidence rate of 1 ASD per 500 children aged eight years, with a higher incidence in boys (23.6 per 1000) when compared to girls (5.3 per 1000).[2] ASD-affected individuals exhibit unusual ways of learning and reactions to sensation. ASD is a multifactorial disorder caused by the interaction of both genetic and non-genetic risk factors.
2. Etiological Factors Leading to Autism
Mounting evidence revealed that de novo mutations, copy number variations, rare and common variants are major genetic factors leading to ASD. Around 50% of ASDs are hereditary caused due to defects in the gene and chromosomal abnormalities leading to disruption in the neuronal connection, brain growth, and synaptic morphology.[3][4][5][6] Siblings born in families with ASD have a 50% enhanced risk of ASD with a reoccurrence rate of 5%–8%. In monozygotic twins, the concordance rate is 90%, while in dizygotic twins the rate of incidence is 10%.[7] Genetic studies revealed that a mutation in the single gene involved in synaptogenesis alters the developmental pathways of neuronal and axonal structures. The fragile X syndrome, tuberous sclerosis, hyperexcitability of neocortical circuits and abnormal neural synchronization were considered as probable disorders leading to ASD.[8][9] In-depth genomic studies revealed that the chromosomes 2q, 7q, 15q, and 16p have genes susceptible for ASD that have not yet been completely studied.[10] Inborn metabolic errors such as phenylketonuria, creatine deficiency syndromes, adenylosuccinate lyase deficiency, and metabolic purine disorders account for 5% of ASD incidence. Recent reports revealed that the gene ENGRAILED 2 mainly involved in cerebellar developmental patterning, GABA system genes, and serotonin transporter genes, has been considered to be associated with ASD.[11]
Non-predisposed factors such as exposure to environmental factors and pharmaceutical drugs, an autoimmune disorder, microbial infection and diet during the prenatal and postnatal periods cause gut dysbiosis and immune dysregulation together contributing to ASD (Figure 1). Recent research revealed that the severity of ASD depends on the complex interaction of genetic susceptibility and environmental factors, so unraveling this relationship will help in identifying a treatment strategy for ASD.[12]
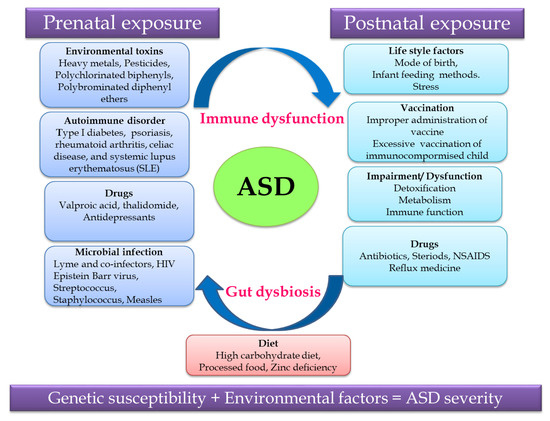
Figure 1. Putative autism spectrum disorder (ASD)-related and environmental factors contributing to ASD.
3. The link between Gut Microbiome and ASD
3.1. Role of Gut Microbiota in Human Nutrition and Health
Gut-brain cross-talk involves a complex communication system involved in the proper maintenance of GI homeostasis, which is termed as the gut-brain axis (GBA). The GBA is a bidirectional communication network between the central nervous system (CNS) and the enteric nervous system linking the emotional and cognitive centers of the brain with peripheral intestinal functions via neuro–immuno–endocrinal mediators.[13] The bidirectional communication network of the GBA involves the brain and spinal cord of the CNS, autonomous nervous system (ANS), the enteric nervous system (ENS) and the hypothalamic pituitary adrenal axis (HPA). Sympathetic and parasympathetic limbs of ANS transmit the afferent signals from the lumen to the CNS through the enteric, spinal and vagal pathways and efferent signals from the CNS to the intestinal wall.[14] The HPA axis is the core stress efferent axis, which adapts the organism to various stresses and is part of the limbic system involved in memory and emotional response. On exposure to stresses such as environmental factors and pro-inflammatory cytokines, the HPA axis activates the release of the corticotrophin release factor (CRF) from the hypothalamus, stimulating the secretion of the adrenocorticotrophic hormone (ACTH) from the pituitary gland, which in turn activates the adrenal gland to release the stress hormone cortisol affecting various organs including the brain. Hence, both neuronal and hormonal interactions play a vital role in influencing the activities of intestinal functional effector cells, such as immune cells, epithelial cells, enteric neurons, smooth muscle cells, interstitial cells of cajal and enterochromaffin cells which in turn are under the control of gut microbiota via brain–gut reciprocal communication.[15] Moreover, the epithelial cell lining of the GI tract and its motility, which is controlled by the CNS, influences the composition of the gut microbiome. Hence, any dysregulation in the CNS alters the intestinal microbiota leading to pathological consequences and gut microbial dysbiosis which affects the development and regulation of the hypothalamic-pituitary-adrenal axis (HPA) and behavior. This bidirectional relationship of the gut microbiome with the host brain axis is termed as microbial endocrinology or inter-kingdom signaling.[16]
Gut microbiota interacts with the brain through endocrine and neurocrine pathways, while the brain influences the microbial composition through the autonomic nervous system with the active involvement of the immune and humoral systems. Gut microbiome modulates the brain by influencing the production of neurotransmitters such as serotonin, gamma aminobutyric acid (GABA) and the brain-derived neurotrophic factor (BDNF) via short-chain fatty acid, tryptophan metabolites, secondary bile acids, and ketones thereby influencing memory and learning processes. These molecules transmit signals by interacting with the farnesoid receptor (FXR) and G protein-coupled bile acid receptor (TGR5) in the enteroendocrine cells (EEC) releasing fibroblast growth factor (FGF19), which readily crosses the blood-brain barrier and regulates the secretion of neuropeptide Y in the hypothalamus, thereby regulating the glucose metabolism via the release of glucagon-like peptide (GLP-1).[17][18][19] The release of serotonin (5 hydroxy tryptamine-5HT) by enterochromaffin cells (ECC), triggered by the stimuli from efferent neurons of the CNS based on the availability of the dietary tryptophan level, which in turn is controlled by the gut microbiome, represents the bidirectional gut microbiome–brain axis.[20] Certain microbially-derived molecules escape the intestinal barrier, reach the brain directly by crossing the blood-brain barrier via systemic circulation and propagate the signal on interaction with the FXR and TGR5 expressed in brain neurons.[21]
In addition, the secretion of the biologically active peptide by enteroendocrine cells, mainly involved in GBA interaction, is controlled by the nutritional level of the microbiota. SCFA acts as the major signaling molecule mediating the gut microbiome–brain communication, via EEC and ECC. The brain influences the microbial population through several stresses: by altering the size and quality of the mucus secretion, by slowing the recovery of the migratory motor complex pattern, by the induction a transient delaying of gastric emptying, and by enhancing the frequency of cecocolonic spike burst activity, which affect the GI transit modulating the nutrient supply to enteric microbiota. Different psychological stressors in adults and newborns modulate the composition and biomass of enteric microbiota.[22] (Figure 2).
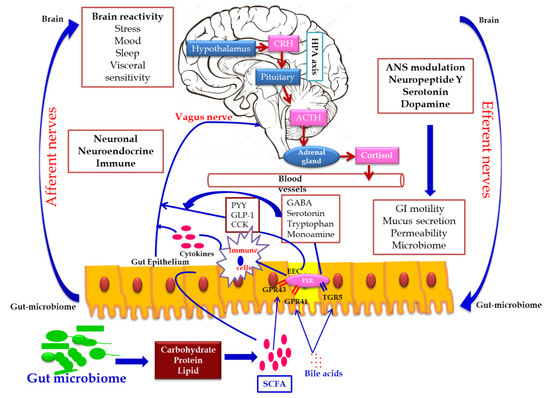
Figure 2. Gut microbiome–brain axis: bidirectional signaling pathways illustrating the relationship between the gut microbiome, intestinal barrier and the brain. Gut microbiota communicates with the brain through the neuro–endocrine–immune network either indirectly via the gut-derived molecules acting on afferent vagal nerve endings, or directly via the microbe-generated signals. The brain’s structural connections (the multiple interconnected structural networks of the central nervous system) regulates the gut microbiota via the autonomic nervous system. Disturbance in the bidirectional interaction response gain is due to psychosocial or gut-derived stress manifests to brain–gut disorders.
3.2. The link between Gut Microbiome and ASD
Gut microbiota, which is non-genetic and inheritable, has a great impact on immune, metabolic and neuronal developments. As gut microbiota is a notable contributor for human health, gut microbial dysbiosis leads to negative consequences such as GI-tract-related disorders such as Crohn’s disease and ulcerative colitis, systemic diseases such as metabolomic disorders and CNS-related disorders.[23] Noticeable evidence illustrated that ASD patients in addition to psychiatric disorders were found to be associated with an extremely painful GI disease termed as autistic enterocolitis or other GI discomforts such as constipation, diarrhea and bloating. This hypothesis leads to the fact that microbial imbalance affects the co-ordination of the microbiota-gut–brain axis in human health leading to several neurological disorders which turned the focus of the researcher towards gut microbiota.
Wakefield and colleagues reported the incidence of a new variant inflammatory bowel disease (autistic enterocolitis), which is characterized by chronic patchy inflammation and lymph nodular hyperplasia in the ileum or colon of individuals with ASD.[24] Scientific evidences also illustrated the relationship between ASD patients and gut microbiome, which has a direct/indirect influence over the feeding pattern and nutrition.[25] The investigation has shown that children with autism suffer from intestinal dysbiosis characterized by the imbalance between beneficial microbes and pathogenic microbes residing in the gut. Neurotoxic and cytotoxic molecules, such as opioid peptides, produced by these pathogenic bacteria enter the bloodstream due to leaky gut, thereby activating the immune mechanism causing tissue damage and GI inflammation. In addition, these toxic molecules affect the neurotransmitter function in the brain, leading to abnormalities in behavioral patterns such as decreased socialization, decreased response to pain, abnormal language, and self-abusive or repetitive behaviors, resulting in confusion, delirium, and even coma.[14] In ASD-vulnerable children, yeast also produces abundant chemicals leading to neurological effects.
The metagenomic analysis of ASD-gut-microbiome showed mucosal dysbiosis with a low level of Bacteriodetes and an increased ratio of Firmicutes to Bacteriodetes. Pyrosequencing of fetal microflora in the fecal stools of autistic children showed a high prevalence of Desulfovibrio species and Bacteroides vulgatus when compared to healthy volunteers.[26] The 16S r DNA sequencing of gut microbiome, isolated from late onset autism patients, showed a high incidence of Clostridium and Ruminococcus species, while a real-time PCR analysis showed a rich source of Clostridium cluster groups I and XI and Clostridium bolteae.[27] Culture-independent fluorescence in situ hybridization studies revealed the elevated level of Clostridium hystolyticum in the ASD children compared to healthy children. [28] The accumulation of neurotoxin-producing bacteria such as Clostridia worsens the autistic symptom. Another study on the gut microbial composition of autistic children revealed a low level of Bifidobacterium and Enterococcus and an increased level of Lactobacillus strains, despite being beneficial, which is quite paradoxical. Commensal bacteria such as Bacillus spp. and Klebsiella oxytoca, that are neither harmful nor beneficial, were reported to be present in autistic children.[29] A pilot study by Kang et al. [30] reported the presence of low-level carbohydrate degrading/fermenting bacteria such as Prevotella, Coprococcus and Veillonellaceae in ASD, substantiating the link between gut microbiome and ASD. Pyrosequencing results showed the altered gut microbial diversity in autistic children with a relatively high abundance of Caloramator, Sarcina and Clostridium genera, Alistipes and Akkermansia species, Sutterellaceae and Enterobacteriaceae and low level of Prevotella, Coprococcus and unclassified Veillonellaceae, concomitantly associated with the altered level of free amino acids and volatile organic compounds in fecal samples compared to the control samples.[31] The above reports illustrate altered gut microbiota in patients suffering from autism, when compared to healthy volunteers, which shows the direct/indirect relationship of gut microbiome with autism.
3.3. Microbial Metabolites Interrelated with ASD
The metabolites produced by gut microbiome affect the neural process based on their level. Metabolomic studies in urine, fecal and serum samples of ASD patients using LC-MS and GC-MS revealed an enhanced level of microbial metabolites, such as increased levels of SCFAs, para-cresol and ammonia, which affect the neural process.[32][33] SCFA is the double edged sword that plays a vital role in human health and disease. SCFA is considered as a major trigger factor for ASD. Propionic acid, the widely used preservative in the food industry, is also one of the SCFAs produced by ASD-associated bacteria, such as Clostridium, Bacteroides and Desulfovibrio. Experimental studies with rodents treated with propionic acid exhibited ASD-associated symptoms, such as impaired and restricted social behavior and cognition, together with an enhanced neuro-inflammatory response, which might be due to alteration in mitochondrial function or the epigenetic modulation of ASD-associated genes.[34] The elevated level of another microbial metabolite para-cresol (p-cresol) and its conjugate p-cresylsulfate were observed in the urinary samples of children affected by autism. The increased level of p-cresol, derived either from the environment or gut microbiome, aggravates the ASD severity by inhibiting the neurotransmitters associated with enzymes and cofactors required for sulfonation reaction in the liver.[35]
Urinary metabolomic research in ASD children revealed the presence of an abnormal level of common microbial metabolites such as dimethylamine, hippuric and phenylacetylglutamine and altered tryptophan, when compared to healthy children.[36][37] The increased level of tyrosine analogue 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), reported in the urine samples of autistic patients, might be responsible for the catecholamine depletion, worsening autistic symptoms such as stereotypical behavior, hyperactivity and hyper-reactivity.[38] Serum metabolomic studies depicted the presence of 11 metabolites in abnormal level among which, the level of sphingosine 1-phosphate and docosahexaenoic acid were consistent in all models.[39] The abnormal gut microbiome-associated metabolites in ASD patients revealed an altered metabolism leading to the aberrant increase in metabolites, worsening the symptoms of ASD.
3.4. Gut Microbiome-Associated Immune Deregulation
Several studies in human and animal models of ASD revealed the presence of an enhanced level of pro-inflammatory cytokines, brain-specific auto-antibodies in the cerebrospinal fluid and serum, illustrating an elevated immune response substantiating the fact that immune dysregulation acts as a key factor contributing to the pathophysiology of ASD.[40][41] Further autopsy of the brain specimens of ASD patients showed the presence of activated microglial cells, together with an increased level of cytokine, such as interferon (IFN)-γ, IL-1β, IL-6, IL-12p40, tumor necrosis factor (TNF)-α and chemokine C-C motif ligand (CCL)-2.[42][43] Elevated cytokines and chemokines were related to aberrant stereotypical behavior and cognitive impairment. In the newborn infants, the immune system development and homeostasis are regulated by the maternal gut microbiome colonizing the fetal gut. Multiple evidences revealed that the immune dysregulation in ASD is associated with gut microbiota[44] Certain species of gut microbiota regulate the T lymphocyte differentiation, while few microbes, such as Bacteroides fragilis colonization, restrict T helper cell response eliciting an autoimmune disorder.[45] Although several studies reported that impairment of the immune system in ASD is linked with gut microbiome, the mechanism behind it is not clearly known.
3.5. Maternal Risk Factors Regulating Gut Microbiome
Epidemiological and experimental studies revealed a strong linkage between maternal infection and the development of ASD in the offspring. The gut microbial composition of a newborn infant varies with respect to the mode of delivery primarily colonized by the maternal microbiota. Hence, any imbalance in maternal microbiome with respect to environmental stress or genetic risk will be transferred to the offspring at the time of birth.[46][47] Scientific evidences revealed that a maternal high-fat diet and exposure to stress during the gestation period increases the risk of neurodevelopment and behavioral disorders in offspring.[48] Maternal immune activation studies using animal models exposed to poly (I:C) during the prenatal stage revealed a change in gut microbiome, leading to lifelong neuropathology and altered behaviors in the offspring.[49][50][51] Based on the reports, it is clear that maternal risk factors increase the incidence of ASD in the offspring by multiple pathways, such as the altered modulation of the placenta, epigenetic modification and immune dysregulation.
References
- Marc Fakhoury; Autistic spectrum disorders: A review of clinical features, theories and diagnosis. International Journal of Developmental Neuroscience 2015, 43, 70-77, 10.1016/j.ijdevneu.2015.04.003.
- Jon Baio; Lisa Wiggins; Deborah L. Christensen; Matthew J Maenner; Julie Daniels; Zachary Warren; Margaret Kurzius-Spencer; Walter Zahorodny; Cordelia Robinson; Cordelia Robinson Rosenberg; et al.Tiffany WhiteMaureen S. DurkinPamela ImmLoizos NikolaouMarshalyn Yeargin-AllsoppLi-Ching LeeRebecca HarringtonMaya LopezRobert T. FitzgeraldAmy HewittSydney PettygroveJohn N. ConstantinoAlison VehornJosephine ShenoudaJennifer Hall-LandeKim VanNaardenKim Van Naarden BraunNicole F. Dowling Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR. Surveillance Summaries 2018, 67, 1-23, 10.15585/mmwr.ss6706a1.
- Luis De La Torre-Ubieta; Hyejung Won; Jason L. Stein; Daniel H Geschwind; Advancing the understanding of autism disease mechanisms through genetics. Nature Medicine 2016, 22, 345-361, 10.1038/nm.4071.
- Stephan J Sanders; Xin He; A. Jeremy Willsey; A. Gulhan Ercan-Sencicek; Kaitlin E. Samocha; A. Ercument Cicek; Michael T. Murtha; Vanessa H. Bal; Somer L. Bishop; Shan Dong; et al.Arthur P. GoldbergCai JinluJohn F. KeaneyLambertus KleiJeffrey MandellDaniel Moreno-De-LucaChristopher S. PoultneyElise RobinsonLouw SmithTor Solli-NowlanMack Y. SuNicole A. TeranMichael F. WalkerDirk WerlingArthur L. BeaudetRita M. CantorEric FombonneDaniel H. GeschwindDorothy E. GriceCatherine LordJennifer K. LoweShrikant M. Manenna M. MartinEric M. MorrowMichael E. TalkowskiJames SutcliffeChristopher A. WalshTimothy W. YuAutism Sequencing ConsortiumDavid H. LedbetterChrista Lese MartinEdwin H. CookJoseph D. BuxbaumMark J. DalyBernie DevlinKathryn RoederMatthew W. State Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci.. Neuron 2015, 87, 1215-1233, 10.1016/j.neuron.2015.09.016.
- Joon-Yong An; Kevin Lin; Lingxue Zhu; Donna M. Werling; Shan Dong; Harrison Brand; Harold Z Wang; Xuefang Zhao; Grace B. Schwartz; Ryan L Collins; et al.Benjamin B. CurrallClaudia DastmalchiJeanselle DeaClif DuhnMichael C. GilsonLambertus KleiLindsay LiangEirene Markenscoff-PapadimitriouSirisha PochareddyNadav AhituvJoseph D. BuxbaumHilary CoonMark J. DalyYoung Shin KimGabor T. MarthBenjamin M. NealeAaron R. QuinlanJohn L. RubensteinNenad SestanMatthew W. StateA. Jeremy WillseyMichael E. TalkowskiBernie DevlinKathryn RoederStephan J Sanders Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 2018, 362, eaat6576, 10.1126/science.aat6576.
- Michael Gandal; Pan Zhang; Evi Hadjimichael; Rebecca L. Walker; Chao Chen; Shuang Liu; Hyejung Won; Harm Van Bakel; Merina Varghese; Yongjun Wang; et al.Annie W. ShiehJillian HaneySepideh ParhamiJudson BelmontMinsoo KimPatricia Moran LosadaZenab KhanJustyna MleczkoYan XiaRujia DaiDaifeng WangYucheng T. YangMin XuKenneth FishPatrick R. HofJonathan WarrellDominic FitzgeraldKevin P. WhiteAndrew E. JaffeMette A. PetersMark GersteinChunyu LiuLilia M. IakouchevaDalila PintoDaniel H GeschwindPsychENCODE Consortium Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127, 10.1126/science.aat8127.
- Joachim F. Hallmayer; Sue Cleveland; Andrea Torres; Jennifer Phillips; Brianne Cohen; Tiffany Torigoe; Janet Miller; Angie Fedele; Jack Collins; Karen Smith; et al.Linda LotspeichLisa A. CroenSally OzonoffClara LajonchereJudith K. GretherNeil Risch Genetic heritability and shared environmental factors among twin pairs with autism.. Archives of General Psychiatry 2011, 68, 1095-102, 10.1001/archgenpsychiatry.2011.76.
- Tomoyuki Takano; Takano T.; Interneuron Dysfunction in Syndromic Autism: Recent Advances. Developmental Neuroscience 2015, 37, 467-475, 10.1159/000434638.
- Maham Rais; Devin K. Binder; Khaleel A. Razak; Iryna M. Ethell; Sensory Processing Phenotypes in Fragile X Syndrome. ASN Neuro 2018, 10, 1759091418801092, 10.1177/1759091418801092.
- Barbara Wiśniowiecka-Kowalnik; Beata Anna Nowakowska; Genetics and epigenetics of autism spectrum disorder—current evidence in the field. Journal of Applied Genetics 2019, 60, 37-47, 10.1007/s13353-018-00480-w.
- Jaume Campistol; María Díez‐Juan; Laura Callejón; Aroa Fernandez‐De Miguel; Mercedes Casado; Angels Garcia Cazorla; Reymundo Lozano; Rafael Artuch; Inborn error metabolic screening in individuals with nonsyndromic autism spectrum disorders. Developmental Medicine & Child Neurology 2016, 58, 842-847, 10.1111/dmcn.13114.
- Varvara Mazina; Jennifer Gerdts; Sandy Trinh; Katy Ankenman; Tracey Ward; Megan Y. Dennis; Santhosh Girirajan; Evan E. Eichler; Raphael Bernier; Epigenetics of autism-related impairment: copy number variation and maternal infection.. Journal of Developmental & Behavioral Pediatrics 2015, 36, 61-7, 10.1097/DBP.0000000000000126.
- Emeran A. Mayer; Kirsten Tillisch; Arpana Gupta; Gut/brain axis and the microbiota.. Journal of Clinical Investigation 2015, 125, 926-38, 10.1172/JCI76304.
- Clair R. Martin; Vadim Osadchiy; Amir Kalani; Emeran A. Mayer; The Brain-Gut-Microbiome Axis. Cellular and Molecular Gastroenterology and Hepatology 2018, 6, 133-148, 10.1016/j.jcmgh.2018.04.003.
- Emeran A. Mayer; Tor Savidge; Robert J. Shulman; Brain-gut microbiome interactions and functional bowel disorders.. Gastroenterology 2014, 146, 1500-12, 10.1053/j.gastro.2014.02.037.
- Hadar Neuman; Justine W Debelius; Rob Knight; Omry Koren; Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiology Reviews 2015, 39, 509-521, 10.1093/femsre/fuu010.
- Hung Hsuchou; Weihong Pan; Abba J Kastin; Fibroblast growth factor 19 entry into brain. Fluids and Barriers of the CNS 2013, 10, 32-32, 10.1186/2045-8118-10-32.
- Genevieve Marcelin; Young-Hwan Jo; Xiaosong Li; Gary J. Schwartz; Ying Zhang; Nae J. Dun; Rong-Ming Lyu; Clémence Blouet; Jaw K. Chang; Streamson C. Chua; et al. Central action of FGF19 reduces hypothalamic AGRP/NPY neuron activity and improves glucose metabolism.. Molecular Metabolism 2013, 3, 19-28, 10.1016/j.molmet.2013.10.002.
- Annika Wahlström; Sama I. Sayin; Hanns-Ulrich Marschall; Fredrik Bäckhed; Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabolism 2016, 24, 41-50, 10.1016/j.cmet.2016.05.005.
- Doe-Young Kim; Michael Camilleri; Serotonin: a mediator of the brain–gut connection. The American Journal of Gastroenterology 2000, 95, 2698-2709, 10.1016/s0002-9270(00)01970-5.
- Mark Klitgaard Nøhr; Kristoffer Lihme Egerod; S.H. Christiansen; Andreas Gille; Stefan Offermanns; Thue W. Schwartz; Morten Møller; Expression of the short chain fatty acid receptor GPR41/FFAR3 in autonomic and somatic sensory ganglia. Neuroscience 2015, 290, 126-137, 10.1016/j.neuroscience.2015.01.040.
- Gil Sharon; Timothy R. Sampson; Daniel H. Geschwind; Sarkis K. Mazmanian; The Central Nervous System and the Gut Microbiome.. Cell 2016, 167, 915-932, 10.1016/j.cell.2016.10.027.
- Walburga Dieterich; Monic Schink; Yurdagül Zopf; Microbiota in the Gastrointestinal Tract. Medical Sciences 2018, 6, 116, 10.3390/medsci6040116.
- Andrew J. Wakefield; J M Puleston; S M Montgomery; A Anthony; J J O'leary; S H Murch; Review article: the concept of entero-colonic encephalopathy, autism and opioid receptor ligands.. Alimentary Pharmacology and Therapeutics 2002, 16, 663-674.
- William G. Sharp; David L. Jaquess; Colleen T. Lukens; Multi-method assessment of feeding problems among children with autism spectrum disorders. Research in Autism Spectrum Disorders 2013, 7, 56-65, 10.1016/j.rasd.2012.07.001.
- Sydney M Finegold; Scot E. Dowd; Viktoria Gontcharova; Chengxu Liu; Kathleen E. Henley; Randall D. Wolcott; Eunseog Youn; Paula H. Summanen; Reen Granpeesheh; Dennis Dixon; et al.Minghsun LiuDenise R. MolitorisJohn A. Green Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444-453, 10.1016/j.anaerobe.2010.06.008.
- Brent L. Williams; Mady Hornig; Tanmay Parekh; W. Ian Lipkin; Application of Novel PCR-Based Methods for Detection, Quantitation, and Phylogenetic Characterization of Sutterella Species in Intestinal Biopsy Samples from Children with Autism and Gastrointestinal Disturbances. mBio 2012, 3, e00261-11, 10.1128/mBio.00261-11.
- Helena Mrt Parracho; Max O Bingham; Glenn R Gibson; Anne L McCartney; Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. Journal of Medical Microbiology 2005, 54, 987-991, 10.1099/jmm.0.46101-0.
- Mingyu Xu; Xuefeng Xu; Jijun Li; Fei Li; Association Between Gut Microbiota and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis.. Frontiers in Psychology 2019, 10, 473, 10.3389/fpsyt.2019.00473.
- Dae-Wook Kang; James B. Adams; Devon M. Coleman; Elena L. Pollard; Juan Maldonado; Sharon McDonough-Means; J. Gregory Caporaso; Rosa Krajmalnik-Brown; Long-term benefit of Microbiota Transfer Therapy on autism symptoms and gut microbiota. Scientific Reports 2019, 9, 5821, 10.1038/s41598-019-42183-0.
- Maria De Angelis; Ruggiero Francavilla; Maria Piccolo; Andrea De Giacomo; Marco Gobbetti; Autism spectrum disorders and intestinal microbiota. Gut Microbes 2015, 6, 207-213, 10.1080/19490976.2015.1035855.
- Derrick F. Macfabe; Short-chain fatty acid fermentation products of the gut microbiome: implications in autism spectrum disorders. Microbial Ecology in Health and Disease 2012, 23, 1, 10.3402/mehd.v23i0.19260.
- Lv Wang; Michael Allan Conlon; Claus T. Christophersen; Michael J. Sorich; Manya Therese Angley; Gastrointestinal microbiota and metabolite biomarkers in children with autism spectrum disorders. Biomarkers in Medicine 2014, 8, 331-344, 10.2217/bmm.14.12.
- Latifa S. Abdelli; Aseela Samsam; Saleh A. Naser; Propionic Acid Induces Gliosis and Neuro-inflammation through Modulation of PTEN/AKT Pathway in Autism Spectrum Disorder.. Scientific Reports 2019, 9, 8824, 10.1038/s41598-019-45348-z.
- Laura Altieri; Cristina Neri; Roberto Sacco; Paolo Curatolo; Arianna Benvenuto; Filippo Muratori; Elisa Santocchi; Carmela Bravaccio; Carlo Lenti; Monica Saccani; et al.Roberto RigardettoMarina GandioneA. UrbaniAntonio M. Persico Urinary p -cresol is elevated in small children with severe autism spectrum disorder. Biomarkers 2011, 16, 252-260, 10.3109/1354750x.2010.548010.
- Ivan K. S. Yap; Manya Angley; Kirill Veselkov; Elaine Holmes; John C. Lindon; Jeremy Nicholson; Urinary Metabolic Phenotyping Differentiates Children with Autism from Their Unaffected Siblings and Age-Matched Controls. Journal of Proteome Research 2010, 9, 2996-3004, 10.1021/pr901188e.
- Federica Gevi; Lello Zolla; Stefano Gabriele; Antonio M. Persico; Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Molecular Autism 2016, 7, 47, 10.1186/s13229-016-0109-5.
- William Shaw; Increased urinary excretion of a 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), an abnormal phenylalanine metabolite ofClostridiaspp. in the gastrointestinal tract, in urine samples from patients with autism and schizophrenia. Nutritional Neuroscience 2010, 13, 135-143, 10.1179/147683010x12611460763968.
- Han Wang; Shuang Liang; Maoqing Wang; Jingquan Gao; Caihong Sun; Jia Wang; Wei Xia; Shiying Wu; Susan Sumner; Fengyu Zhang; et al.Chang-Hao SunLijie Wu Potential serum biomarkers from a metabolomics study of autism. Journal of Psychiatry and Neuroscience 2016, 41, 27-37, 10.1503/jpn.140009.
- Elaine Y. Hsiao; Immune Dysregulation in Autism Spectrum Disorder. International Review of Neurobiology 2013, 113, 269-302, 10.1016/b978-0-12-418700-9.00009-5.
- Geir Bjørklund; Khaled Saad; Salvatore Chirumbolo; Janet K. Kern; David A. Geier; M.R. Geier; Mauricio A. Urbina; Immune dysfunction and neuroinflammation in autism spectrum disorder. Acta Neurobiologiae Experimentalis 2016, 76, 257-268, 10.21307/ane-2017-025.
- Christopher J. Machado; Alexander M. Whitaker; Stephen E.P. Smith; Paul H. Patterson; Melissa D Bauman; Maternal immune activation in nonhuman primates alters social attention in juvenile offspring.. Biological Psychiatry 2014, 77, 823-32, 10.1016/j.biopsych.2014.07.035.
- Jung Won Kim; Ji Yeon Hong; Seung Min Bae; Microglia and Autism Spectrum Disorder: Overview of Current Evidence and Novel Immunomodulatory Treatment Options. Clinical Psychopharmacology and Neuroscience 2018, 16, 246-252, 10.9758/cpn.2018.16.3.246.
- Hsin-Jung Wu; Eric Wu; The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4-14, 10.4161/gmic.19320.
- Hsin-Jung Wu; Ivaylo I. Ivanov; Jaime Darce; Kimie Hattori; Tatsuichiro Shima; Yoshinori Umesaki; Dan R. Littman; Christophe Benoist; Diane Mathis; Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity 2010, 32, 815-27, 10.1016/j.immuni.2010.06.001.
- Paul H. Patterson; Maternal infection and immune involvement in autism. Trends in Molecular Medicine 2011, 17, 389-94, 10.1016/j.molmed.2011.03.001.
- Brian K. Lee; Cecilia Magnusson; Renee M. Gardner; Åsa Blomström; Craig J. Newschaffer; Igor Burstyn; Håkan Karlsson; Christina Dalman; Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain, Behavior, and Immunity 2015, 44, 100-105, 10.1016/j.bbi.2014.09.001.
- Elinor L Sullivan; Elizabeth K Nousen; Katherine A Chamlou; Kevin L Grove; The impact of maternal high-fat diet consumption on neural development and behavior of offspring. International Journal of Obesity Supplements 2012, 2, S7-S13, 10.1038/ijosup.2012.15.
- Mili Mandal; Robert Donnelly; Stella Elkabes; Pan Zhang; Dan Davini; Brian T. David; Nicholas M. Ponzio; Maternal immune stimulation during pregnancy shapes the immunological phenotype of offspring. Brain, Behavior, and Immunity 2013, 33, 33-45, 10.1016/j.bbi.2013.04.012.
- Myka L. Estes; A. Kimberley McAllister; Maternal immune activation: Implications for neuropsychiatric disorders. Science 2016, 353, 772-777, 10.1126/science.aag3194.
- Stefano Nardone; Evan Elliott; The Interaction between the Immune System and Epigenetics in the Etiology of Autism Spectrum Disorders. Frontiers in Neuroscience 2016, 10, 732, 10.3389/fnins.2016.00329.
