Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Laxmi Dongur and Version 2 by Dean Liu.
Donor derived cell-free DNA has been identified as a measurable lab test that may be able to adequately diagnose rejection at early stages, precluding the need for invasive procedures like biopsy.
- renal transplantation
- T-cell mediated rejection
- antibody-mediated rejection
- donor-derived cell-free DNA
1. Introduction
Clinical kidney transplantation has evolved from a niche operation that was historically only possible between identical twins at a few highly specialized centers to a standard of care for patients with end-stage renal disease that can be conducted between donor–recipient pairs from disparate ethnicities and geographies. As the complexities of donor–recipient matching increase alongside theour understanding of the highly variable human immune system, more complex methods of allograft monitoring are needed beyond standard measures of serum creatine and proteinuria. Indeed, despite the routine use of induction and multi-drug maintenance immunosuppression, expected incidences of acute and subclinical rejection can be up to 20–30% and these data come from biopsy-proven pathology [1]. The quest for specific and reproducible noninvasive markers of graft injury has naturally led to genetic markers including donor-derived cell-free DNA (ddcfDNA), released during injury and rejection of an allograft (Figure 1). Here, rwesearchers describe some of the historical advances leading to ddcfDNA as a clinical tool to aid in the diagnosis of rejection in kidney transplantation, review the current uses and limitations, and explore some of the future directions of this exciting biomarker.
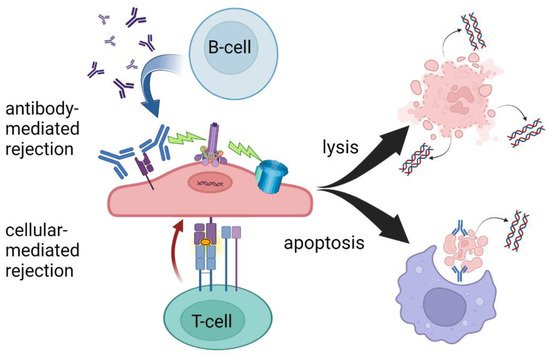
Figure 1. T-cell- and antibody-mediated rejection leading to cell lysis and/or apoptosis. While there is significant overlap in the mechanisms of AMR and TCMR, ddcfDNA levels may be more sensitive to AMR due to increased circulating cellular debris following cell lysis. Created with BioRender.com.
2. History of ddcfDNA in Kidney Transplantation
Although the permanent admixture of donor cellular material into the recipient system in organ transplantation was indirectly suspected as early as 1963 as playing a role in rejection and tolerance, the first direct evidence of DNA micro-chimerism in solid organ transplantation came in 1993 by Starzl et al. [2][3][2,3]. This was made feasible by developing methods to measure the presence of the genetic material of chromosomes 6 (Human Leukocyte Antigen, HLA) and Y (male sex), detecting mismatched donor-derived cells distant from the transplanted organ. The transition from cell-based DNA interrogation (probe-based cytostaining and cellular homogenates) to cell-free DNA detection was made in 1998 by Lo and colleagues when they were able to isolate Y chromosome genetic material from the circulating plasma of female transplant recipients [4]. Despite the limited applicability of the approach (only useful in female recipients of organs from male donors), this represented the first donor-specific cell-free DNA (ddcfDNA) detection in solid organ transplantation and an early demonstration that reliable assumptions about donor genotype could be useful. Further advances in genomic sequencing led to sex-independent methods of distinguishing between the donor and recipient DNA that initially focused on HLA-specific quantitative PCR. Although this approach characterized a significant advance in amplifying very low levels of circulating ddcfDNA, the reproducibility was suboptimal, and the technique suffered from the limitation of not being able to distinguish well between HLA-matched recipient–donor pairs [5]. In 2011, Snyder et al. from Stanford University published a very reliable method for detecting ddcfDNA wherein microfluidic digital PCR was utilized to detect differences in single nucleotide polymorphism (SNP) frequencies between donor and recipient. This method was extremely accurate; however, required full genome sequencing of both the donor and the recipient, a resource and time-intensive endeavor [6]. Nonetheless, in ddthis study, ddcfDNA was significantly correlated with acute rejection in heart transplant recipients and developed the backbone for all commercially available ddcfDNA tests today.
The next major advance toward ddcfDNA becoming a clinically useful tool in organ transplantation came from advances in computational genetics and the developing understating of population-level allelic frequencies stemming from the Human Genome Project [7][8][7,8]. In 2016, Sharon et al. showed that useful measures of ddcfDNA could be obtained in the absence of full genotyping of the donor by analyzing 150–600 thousand SNPs. This method utilized full genotyping of the recipient and was able to distinguish between related and unrelated donor–recipient pairs utilizing the principles of allelic equilibrium and chromosomal inheritance [9]. That same year, Grskovic et al. showed the feasibility of quantifying ddcfDNA with neither a full recipient nor a full donor genotype by interrogating just 266 SNPs [10]. These polymorphisms were targeted due to an extremely low probability of two unrelated individuals having identical genotypes and a low linkage state. Although this method performed equally well in closely and distantly related pairings, it was limited in its ability to identify the presence of more than two distinct genomes as might be found in recipients of transplants from multiple donors. Furthermore, as in all DNA testing, there remained an inability to detect ddcfDNA from monozygotic (identical) twins.
From these myriad advancements, rwesearchers are are able today to utilize ddcfDNA as a practical clinical assay, available via several commercial preparations and approved by many insurance plans to aid in the diagnosis of allograft rejection. Nonetheless, limitations persist in distinguishing among etiologies of allograft injury and theour understanding of the optimal use of these powerful tests is constantly evolving.
3. Overview of Commercially Available ddcfDNA Tests
Today in the United States, three commercially available ddcfDNA assays are available for clinical use in kidney transplantation, Allosure by CareDx, Prospera by Natera, and TRAC by Viracor Eurofins [11][12][13][11,12,13]. These tests all currently require whole blood to be analyzed in a centralized laboratory, a “send out” test, and differ mainly in the number of SNPs measured. At the time of development of these assays, the gold standard for diagnosing rejection was the interpretation of histopathology by a trained pathologist according to the Banff criteria and therefore, all tests were initially validated based on biopsy-proven rejection via classic histology findings [14]. The ubiquitous finding, regardless of platform, that elevated ddcfDNA better predicts antibody-mediated rejection (AMR) compared to T-cell-mediated rejection is a yet poorly understood phenomenon, see Table 1. At the validated thresholds, ddcfDNA is more sensitive to antibody-mediated rejection. This may be due to complement-activated recruitment of the membrane attack complex leading to cell lysis and hence the release of more intracellular debris including cfDNA. The targeting of the microvasculature endothelium may also create an ischemic environment contributing to necrosis. In the setting of T-cell-mediated rejection, phagocytosis following apoptosis may sequester more intracellular contents, leading to less measurable cfDNA despite the presence of graft injury (Figure 1).
Table 1. Summary of characteristics of commercially available clinical ddcfDNA tests in the US for use in transplant recipients with suspicion of acute rejection.
| AlloSure (CareDx) | Prospera (Natera) | TRAC (Viracor Eurofins) | |
|---|---|---|---|
| Initial Validation Cohort | DART (NCT01299168, 2015–2016) [15] prospective 14 sites 102 patients |
UCSF biobank (pre-2018) * [16] retrospective 1 site 178 patients |
Undisclosed biobank (pre-2020) ** [17] retrospective 1 site 25 patients |
| Calibration Standard | Histopathology (BPR) | Histopathology (BPR) | Histopathology (BPR) |
| Targeted diagnosis | Acute rejection (AMR > TCR) | Acute rejection (AMR > TCR) | Acute rejection (AMR > TCR) |
| Suggested threshold | 1% | 1% | 0.7% |
| Reported sensitivity, specificity | 59%, 85% | 89%, 73% | 58%, 85% |
| NPV, PPV *** | 84%, 61% | 95%, 52% | 86%, 55% |
| Potential false positive rate *** | 15% | 27% | 15% |
DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study; UCSF, University of California San Francisco; BPR, biopsy-proven rejection; AMR, antibody-mediated rejection; TCR, T-cell-mediated rejection; NPV, negative predictive value; PPV, positive predictive value. * Study duration not included in landmark publication, subsequently studied in TRIFECTA (NCT04239703, 2019-recruiting, n = 367). ** initially studied in R&D setting, subsequently studied post hoc via CTOT-08 (NCT01289717, 2011-2016) and single center biobank (Northwestern University). *** at 25% prevalence.
4. Current Uses
The tests have evolved to be employed in conjunction with tests such as circulating leukocyte gene expression markers due in part to the fact that they all share relatively high negative predictive values (NPV) and relatively low positive predictive values (PPV). In short, the absence of ddcfDNA is a better predictor of the state of an allograft, in terms of rejection, than the presence of ddcfDNA, which may represent driving forces other than rejection. Outside of the landmark original validation studies, this finding has been reproduced by several groups, with NPV being consistently higher than PPV [18][19][20][21][22][18,19,20,21,22]. Additionally, the clinical performance of commercially available ddcfDNA tests appears to be similar (commonly used cutoff value of ~1%) despite different validation strategies [23][24][23,24]. Therefore, the main utility in the clinical setting, consistent with the original intention, is to confidently rule out suspected rejection and avoid a potentially unnecessary biopsy. Indeed, with the rare but potentially catastrophic complications of percutaneous biopsy combined with the inherent diagnostic limitations, avoiding even one biopsy in the lifetime of a kidney recipient represents a substantial improvement [25].
