Despite the advancements in cancer treatments, gastric cancer is still one of the leading causes of death worldwide. In this context, it is of great interest to discover new and more effective ways of treating this disease. Accumulated evidences have demonstrated the amplification of 8q24.21 region in gastric tumors. Furthermore, this is the region where the widely known MYC oncogene and different microRNAs are located. MYC deregulation is key in tumorigenesis in various types of tissues, once it is associated with cell proliferation, survival, and drug resistance. microRNAs are a class of noncoding RNAs that negatively regulate the protein translation, and which deregulation is related with gastric cancer development. However, little is understood about the interactions between microRNAs and MYC. Here, we overview the MYC role and its relationship with the microRNAs network in gastric cancer aiming to identify potential targets useful to be used in clinic, not only as biomarkers, but also as molecules for development of promising therapies.
- gastric cancer
- microRNA
- 8q24.21
- MYC
- target therapy
1. Introduction
The MYC family is a group of cellular proto-oncogenes with the following three highly related nuclear phosphoproteins: MYC, N-MYC, and L-MYC[1] [61]. MYC has a low expression and has a short half-life in normal cells, and its mRNA level is tightly regulated by both transcriptional and post-transcriptional mechanisms[2] [62]. However, it is overexpressed in several neoplasms.
2. Biological Significance of MYC
Our group and others have shown MYC overexpression in GC samples[3][4][5] [17,43,44], including early stages[6][7] [23,63], and reported MYC protein overexpression[6][8] [24,23]. Moreover, other studies revealed the importance of the co-amplification of MYC and EGFR and FGFR2, in predicting poor survival of patients undergoing cancer therapy[9] [64]. In tumor cells, MYC activation occurs as follows: (1) mutations in signaling pathways proteins upstream from MYC; (2) mutations and single nucleotide polymorphisms in regulatory regions that enhance the stability of these proteins[10] [65] and (3) direct modification of MYC gene via gene amplification, mutation, chromosomal translocation and epigenetic modifications[6][10][11][12] [24,65–67].
MYC deregulation plays an important role in neoplastic development by targeting genes involved in critical cellular functions, such as DNA metabolism and dynamics, cell cycle, apoptosis, adhesion, survival, and protein and macromolecular synthesis[2][13][14] [62,68,69]. Moreover, it contributes to aerobic metabolism by activating the expression of several genes essential for glycolysis and mitochondrial biogenesis[15] [70]. Additionally, its hyperactivity can allow widespread miRNAs downregulation through the regulation of transcriptional and post-transcriptional mechanisms. Indeed, MYC is known as the gene with the highest interaction with downregulated miRNAs[16][17][18] [71–73]. Taken together, this scenario shows that MYC deregulation (usually overexpression) can have an impact in various cellular functions, contributing to an abnormal cell growth (Figure 1)[15][19] [70,74].
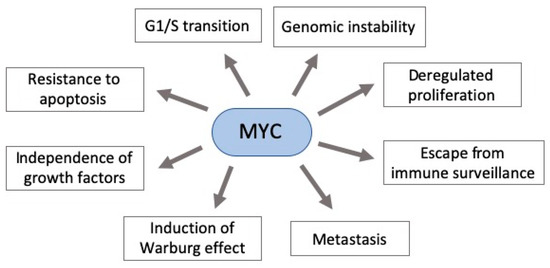
Figure 1. Pleiotropic consequence of MYC deregulation in cancer. MYC overexpression in gastric carcinogenesis affects various components of signaling pathways critical to cancer establishment. Some of these pathways’ phenotypes are shown here.
Infectious agents are extremely important factors on cancer development, accounting for 16% of all new cancer cases per year worldwide[20] [75]. Moreover, liver and gastric tumors in men account for greater than 80% of the infection-related burden cancers[20] [75]. According to the International Agency for Research on Cancer (IARC), 78% of all GC cases are estimated to be associated to chronic H. pylori infection, a bacteria classified as a group 1 carcinogen[21] [76]. The virulence of this bacterium is commonly determined by cagA and vacA genes. The cagA gene encodes the secretion complex, capable of introducing the cagA oncoprotein in the gastric epithelial cell, which activates mitogen-activated protein (MAP) kinases. This alteration activates cell proliferation, differentiation, and stress and inflammatory responses and inhibits programed death, leading to a precancerous process[22][23] [77,78]. Especially in intestinal-type of GC, H. pylori cagA has been associated with increased MYC expression and nuclear MYC protein[24][25] [79,80]. In H. pylori infected patients with active gastritis, chromosomal aneuploidy and cellular DNA damage were associated with MYC expression, leading to a chronic hyperproliferation[26] [81]. This association may occur through H. pylori-induced activation of NF-κB and AP-1 proteins which transcriptionally regulate β-catenin expression, responsible for controlling MYC expression and consequently cell proliferation[14][27] [69,82]. On the other hand, MYC overexpression was not observed in patients without H. pylori infection[26] [81].
The alteration of the DNA methylation profile is considered to be associated with the H pylori inflammatory response, rather than the infection itself[28] [83]. This infection participates in the regulation of MYC expression, which is necessary to gastric carcinogenesis occur (Figure 2), but its infection alone is insufficient to the disease establishment. Thus, the identification of molecules and miRNAs associated with H. pylori infection in GC can contribute to understand the key cellular and molecular processes at the beginning of carcinogenesis and how environmental factors contribute to GC etiology.
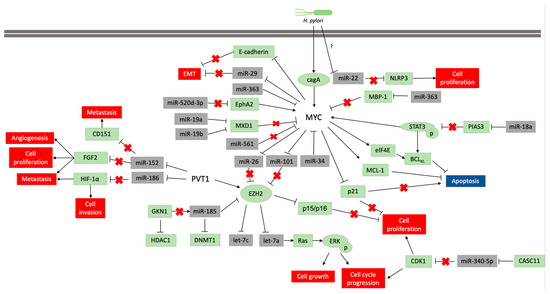
Figure 2: Pathways scheme in which miRNAs regulate MYC and PVT1 expression and vice versa in neoplastic gastric tissue samples and cell lines. The coding genes are shown in green, and the non-coding genes in grey. Lines ending with an arrow indicate activation, whereas T ending lines indicate repression. Lines with a red cross indicate that the interaction is lost due to repression or blocking of a miRNA or protein.
