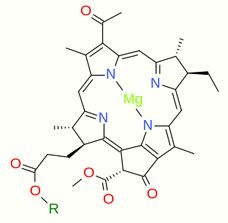
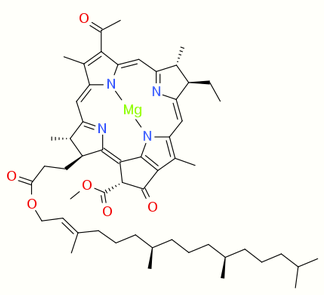
Chlorophyllide a and Chlorophyllide b are the biosynthetic precursors of chlorophyll a and chlorophyll b respectively. Their propionic acid groups are converted to phytyl esters by the enzyme chlorophyll synthase in the final step of the pathway. Thus the main interest in these chemical compounds has been in the study of chlorophyll biosynthesis in plants, algae and cyanobacteria. Chlorophyllide a is also an intermediate in the biosynthesis of bacteriochlorophylls.
- chlorophyll biosynthesis
- chlorophyll b
- chlorophyll a
1. Structures
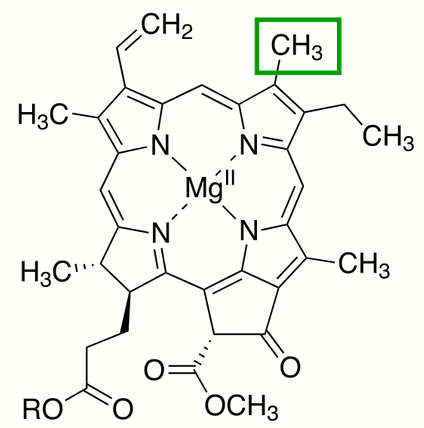
Chlorophyllide a, is a carboxylic acid (R=H). In chlorophyllide b, the methyl group at position 13 (IUPAC numbering for chlorophyllide a) and highlighted in the green box, is replaced with a formyl group.
2. Biosynthesis Steps up to Formation of Protoporphyrin IX
In the early steps of the biosynthesis, which starts from glutamic acid, a tetrapyrrole is created by the enzymes deaminase and cosynthetase which transform aminolevulinic acid via porphobilinogen and hydroxymethylbilane to uroporphyrinogen III. The latter is the first macrocyclic intermediate common to haem, sirohaem, cofactor F430, cobalamin and chlorophyll itself.[1] The next intermediates are coproporphyrinogen III and protoporphyrinogen IX, which is oxidised to the fully aromatic protoporphyrin IX. Insertion of iron into protoporphyrin IX in for example mammals gives haem, the oxygen-carrying cofactor in blood, but plants combine magnesium instead to give, after further transformations, chlorophyll for photosynthesis.[2]
3. Biosynthesis of Chlorophyllides from Protoporphyrin IX
Details of the late stages of the biosynthetic pathway to chlorophyll differ in the plants (for example Arabidopsis thaliana, Nicotiana tabacum and Triticum aestivum) and bacteria (for example Rubrivivax gelatinosus and Synechocystis) in which it has been studied. However, although the genes and enzymes vary, the chemical reactions involved are identical.[3][4]
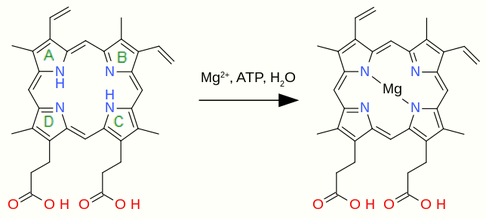
3.1. Insertion of Magnesium
Chlorophyll is characterised by having a magnesium ion coordinated within a ligand called a chlorin. The metal is inserted into protoporphyrin IX by the enzyme magnesium chelatase[3] which catalyzes the reaction EC 6.6.1.1
- protoporphyrin IX + Mg2+ + ATP + H2O [math]\displaystyle{ \rightleftharpoons }[/math] ADP + phosphate + Mg-protoporphyrin IX + 2 H+
3.2. Esterification of the Ring C Propionate Group
The next step towards the chlorophyllides is the formation of a methyl (CH3) ester on one of the propionate groups, which is catalysed by the enzyme Magnesium protoporphyrin IX methyltransferase[5] in the methylation reaction EC 2.1.1.11
- Mg-protoporphyrin IX + S-adenosylmethionine [math]\displaystyle{ \rightleftharpoons }[/math] Mg-protoporphyrin IX 13-methyl ester + S-adenosyl-L-homocysteine
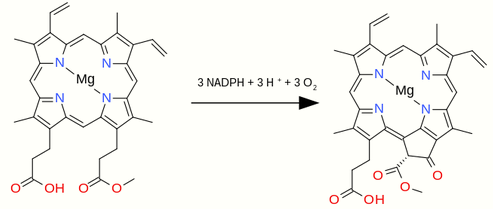
3.3. From Porphyrin to Chlorin
The chlorin ring system features a five-membered carbon ring E is created when one of the propionate groups of the porphyrin is cyclised to the carbon atom linking the original pyrrole rings C and D. A series of chemical steps catalysed by the enzyme Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase[6] gives the overall reaction EC 1.14.13.81
- Mg-protoporphyrin IX 13-monomethyl ester + 3 NADPH + 3 H+ + 3 O2 [math]\displaystyle{ \rightleftharpoons }[/math] divinylprotochlorophyllide + 3 NADP+ + 5 H2O
In barley the electrons are provided by reduced ferredoxin, which can obtain them from photosystem I or, in the dark, from Ferredoxin—NADP(+) reductase: the cyclase protein is named XanL and is encoded by the Xantha-l gene.[7] In anaerobic organisms such as Rhodobacter sphaeroides the same overall transformation occurs but the oxygen incorporated into magnesium-protoporphyrin IX 13-monomethyl ester comes from water in the reaction EC 1.21.98.3.[8]
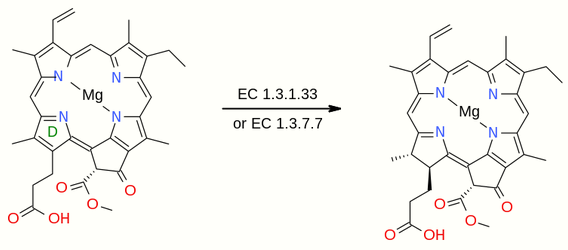
3.4. Reduction Steps to Chlorophyllide a
Two further transformations are required to produce chlorophyllide a. Both are reduction reactions: one converts a vinyl group to an ethyl group and the second adds two atoms of hydrogen to the pyrrole ring D, although the overall aromaticity of the macrocycle is retained. These reactions proceed independently and in some organisms the sequence is reversed.[3] The enzyme divinyl chlorophyllide a 8-vinyl-reductase[9] converts 3,8-divinylprotochlorophyllide to protochlorophyllide in reaction EC 1.3.1.75
- 3,8-divinylprotochlorophyllide + NADPH + H+ [math]\displaystyle{ \rightleftharpoons }[/math] protochlorophyllide + NADP+
This is followed by the reaction EC 1.3.1.33 in which the pyrrole ring D is reduced by the enzyme protochlorophyllide reductase[10]
- protochlorophyllide + NADPH + H+ [math]\displaystyle{ \rightleftharpoons }[/math] chlorophyllide a + NADP+
This reaction is light-dependent but there is an alternative enzyme, Ferredoxin,[11] that uses reduced ferredoxin as its cofactor and is not dependent on light; it performs the a similar reaction EC 1.3.7.7 but with the alternative substrate 3,8-divinylprotochlorophyllide
- 3,8-divinylprotochlorophyllide + reduced ferredoxin + 2 ATP + 2 H2O [math]\displaystyle{ \rightleftharpoons }[/math] 3,8-divinylchlorophyllide a + oxidized ferredoxin + 2 ADP + 2 phosphate
In the organisms which use this alternative sequence of reduction steps, the process is completed by the reaction EC 1.3.7.13 catalysed by an enzyme which can take a variety of substrates and perform the required vinyl-group reduction, for example in this case
- 3,8-divinylchlorophyllide a + 2 reduced ferredoxin + 2 H+ [math]\displaystyle{ \rightleftharpoons }[/math] chlorophyllide a + 2 oxidized ferredoxin
3.5. From Chlorophyllide a to Chlorophyllide b
Chlorophyllide a oxygenase is the enzyme that converts chlorophyllide a to chlorophyllide b[12] by catalysing the overall reaction EC 1.3.7.13
- chlorophyllide a + 2 O2 + 2 NADPH + 2 H+ [math]\displaystyle{ \rightleftharpoons }[/math] chlorophyllide b + 3 H2O + 2 NADP+
4. Use in the Biosynthesis of Chlorophyll a and Chlorophyll b
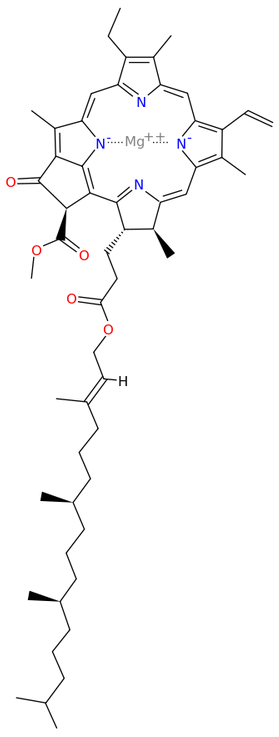
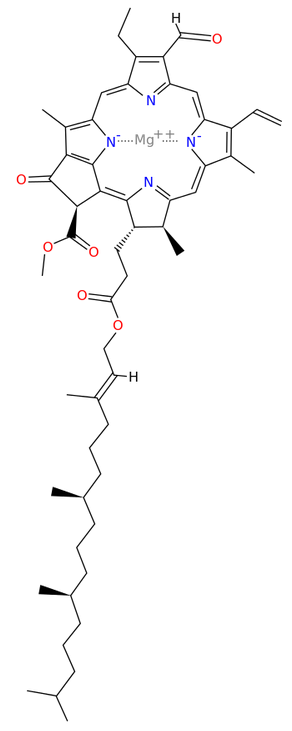
Chlorophyll synthase[13] completes the biosynthesis of chlorophyll a by catalysing the reaction EC 2.5.1.62
- chlorophyllide a + phytyl diphosphate [math]\displaystyle{ \rightleftharpoons }[/math] chlorophyll a + diphosphate
This forms an ester of the carboxylic acid group in chlorophyllide a with the 20-carbon diterpene alcohol phytol. Chlorophyll b is made by the same enzyme acting on chlorophyllide b.
5. Use in the Biosynthesis of Bacteriochlorophylls
Bacteriochlorophylls are the light harvesting pigments found in photosynthetic bacteria: they do not produce oxygen as a side-product. There are many such structures but all are biosynthetically related by being derived from chlorophyllide a.[3][14] Bacteriochlorophyll a is a typical example; its biosynthesis has been studied in Rhodobacter capsulatus and Rhodobacter sphaeroides.
The first step is the reduction (with trans stereochemistry) of the pyrrole ring B, giving the characteristic 18-electron aromatic system of many bacteriochlorophylls. This is carried out by the enzyme chlorophyllide a reductase, which catalyses the reaction EC 1.3.7.15.
- chlorophyllide a + 2 reduced ferredoxin + ATP + H2O + 2 H+ [math]\displaystyle{ \rightleftharpoons }[/math] 3-deacetyl 3-vinylbacteriochlorophyllide a + 2 oxidized ferredoxin + ADP + phosphate
The next two steps convert the vinyl group first into a 1-hydroxyethyl group and then into the acetyl group of bacteriochlorophyllide a. The reactions are catalysed by chlorophyllide a 31-hydratase (EC 4.2.1.165) and bacteriochlorophyllide a dehydrogenase (EC 1.1.1.396) as follows:[15][16]
- 3-deacetyl 3-vinylbacteriochlorophyllide a + H2O [math]\displaystyle{ \rightleftharpoons }[/math] 3-deacetyl 3-(1-hydroxyethyl)bacteriochlorophyllide a
- 3-deacetyl 3-(1-hydroxyethyl)bacteriochlorophyllide a + NAD+ [math]\displaystyle{ \rightleftharpoons }[/math] bacteriochlorophyllide a + NADH + H+
These three enzyme-catalysed reactions can occur in different sequences to produce bacteriochlorophyllide a ready for esterification to the final pigments for photosynthesis. The phytyl ester of bacteriochlorophyll a is not attached directly: rather, the initial intermediate is the ester with R=geranylgeranyl (from geranylgeranyl pyrophosphate) which is then subject to additional steps as three of the sidechain's alkene bonds are reduced.[16]
References
- "Biosynthesis of the pigments of life: formation of the macrocycle". Nature 285 (5759): 17–21. May 1980. doi:10.1038/285017a0. PMID 6769048. Bibcode: 1980Natur.285...17B. https://dx.doi.org/10.1038%2F285017a0
- Battersby, A. R. (2000). "Tetrapyrroles: the Pigments of Life. A Millennium review". Natural Product Reports 17 (6): 507–526. doi:10.1039/B002635M. PMID 11152419. https://dx.doi.org/10.1039%2FB002635M
- Willows, Robert D. (2003). "Biosynthesis of chlorophylls from protoporphyrin IX". Natural Product Reports 20 (6): 327–341. doi:10.1039/B110549N. PMID 12828371. https://dx.doi.org/10.1039%2FB110549N
- R. Caspi (2007-07-18). "3,8-divinyl-chlorophyllide a biosynthesis I (aerobic, light-dependent)". MetaCyc Metabolic Pathway Database. https://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=CHLOROPHYLL-SYN.
- Shepherd, Mark; Reid, James D.; Hunter, C. Neil (2003). "Purification and kinetic characterization of the magnesium protoporphyrin IX methyltransferase from Synechocystis PCC6803". Biochemical Journal 371 (2): 351–360. doi:10.1042/BJ20021394. PMID 12489983. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1223276
- "The Chlorophyll Biosynthetic Enzyme Mg-Protoporphyrin IX Monomethyl Ester (Oxidative) Cyclase (Characterization and Partial Purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803)". Plant Physiology 112 (1): 105–114. September 1996. doi:10.1104/pp.112.1.105. PMID 12226378. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=157929
- Stuart, David; Sandström, Malin; Youssef, Helmy M.; Zakhrabekova, Shakhira; Jensen, Poul Erik; Bollivar, David W.; Hansson, Mats (2020-09-08). "Aerobic Barley Mg-protoporphyrin IX Monomethyl Ester Cyclase is Powered by Electrons from Ferredoxin" (in en). Plants 9 (9): 1157. doi:10.3390/plants9091157. PMID 32911631. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=7570240
- Porra, Robert J.; Schafer, Wolfram; Gad'On, Nasr; Katheder, Ingrid; Drews, Gerhart; Scheer, Hugo (1996). "Origin of the Two Carbonyl Oxygens of Bacteriochlorophyll a. Demonstration of two Different Pathways for the Formation of Ring e in Rhodobacter sphaeroides and Roseobacter denitrificans, and a Common Hydratase Mechanism for 3-acetyl Group Formation". European Journal of Biochemistry 239 (1): 85–92. doi:10.1111/j.1432-1033.1996.0085u.x. PMID 8706723. https://dx.doi.org/10.1111%2Fj.1432-1033.1996.0085u.x
- Parham, Ramin; Rebeiz, Constantin A. (1992). "Chloroplast biogenesis: [4-vinyl] chlorophyllide a reductase is a divinylchlorophyllide a-specific, NADPH-dependent enzyme". Biochemistry 31 (36): 8460–8464. doi:10.1021/bi00151a011. PMID 1390630. https://dx.doi.org/10.1021%2Fbi00151a011
- Apel, Klaus; Santel, Hans-Joachim; Redlinger, Tom E.; Falk, Heinz (2005). "The Protochlorophyllide Holochrome of Barley (Hordeum vulgare L.)". European Journal of Biochemistry 111 (1): 251–258. doi:10.1111/j.1432-1033.1980.tb06100.x. PMID 7439188. https://dx.doi.org/10.1111%2Fj.1432-1033.1980.tb06100.x
- "X-ray crystal structure of the light-independent protochlorophyllide reductase". Nature 465 (7294): 110–4. May 2010. doi:10.1038/nature08950. PMID 20400946. Bibcode: 2010Natur.465..110M. https://dx.doi.org/10.1038%2Fnature08950
- Eggink, Laura L.; Lobrutto, Russell; Brune, Daniel C.; Brusslan, Judy; Yamasato, Akihiro; Tanaka, Ayumi; Hoober, J Kenneth (2004). "Synthesis of chlorophyll b: Localization of chlorophyllide a oxygenase and discovery of a stable radical in the catalytic subunit". BMC Plant Biology 4: 5. doi:10.1186/1471-2229-4-5. PMID 15086960. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=406501
- Schmid, H. C.; Rassadina, V.; Oster, U.; Schoch, S.; Rüdiger, W. (2002). "Pre-Loading of Chlorophyll Synthase with Tetraprenyl Diphosphate is an Obligatory Step in Chlorophyll Biosynthesis". Biological Chemistry 383 (11): 1769–78. doi:10.1515/BC.2002.198. PMID 12530542. https://epub.ub.uni-muenchen.de/17847/1/bc.2002.198.pdf.
- Senge, Mathias O.; Smith, Kevin M. (2004). "Biosynthesis and Structures of the Bacteriochlorophylls". Anoxygenic Photosynthetic Bacteria. Advances in Photosynthesis and Respiration. 2. pp. 137–151. doi:10.1007/0-306-47954-0_8. ISBN 0-7923-3681-X. https://dx.doi.org/10.1007%2F0-306-47954-0_8
- Bollivar, David W. (2007). "Recent advances in chlorophyll biosynthesis". Photosynthesis Research 90 (2): 173–194. doi:10.1007/s11120-006-9076-6. PMID 17370354. https://dx.doi.org/10.1007%2Fs11120-006-9076-6
- R. Caspi (2015-12-08). "Pathway: bacteriochlorophyll a biosynthesis". MetaCyc Metabolic Pathway Database. https://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-5526.