ObesEffity associated with a Western diet such as a high-fat diet (HFD) is a known risk factor for inflammatory bowel cacy evaluation through a disease (IBD) and colorectal cancer (CRC). In this study, we aimed to develop fecal model using the gut microbiome data-based deep learning algorithms for the risk assessment of colorectal dis-eases. The effects of a HFD and a candidate food (Nypa fruticans, NF) on IBD and CRC risk reduction were also evaluated. Fecal microbiome data were obtained from 109 IBD patients, 111 CRC patients, and 395 healthy control (HC) subjects by 16S rDNA amplicon sequencing. IBD and CRC risk as-sessment of well-controlled obese rats using a risk prediction models were then constructed by deep learning algorithms. Dietary effects were evaluated based on fecal based on clinical microbiome data from rats fed on a regular chow diet (RCD), HFD, and HFD plus ethanol extracts or water extracts of NF. There were significant differences in taxa when IBD and CRC were compared with HC. The diagnostic performance (area under curve, AUC) of the deep learning algorithm was 0.84 for IBD and 0.80 for CRC prediction. Based on the rat fecal microbiome data, IBD and CRC risks were increased in HFD-fed rats versus RCD-fed rats. Inter-estingly, in the HFD-induced obesity model, the IBD and CRC risk scores were significantly lowered by the administration of ethanol extracts of NF, but not by the administration of water extracts of NF. In conclusion, changes in the fecal microbiome of obesity by Western diet could be important risk factors for the development of IBD and CRC. The risk prediction model developed in this study could be used to evaluate dietary efficacyor inflammatory bowel disease and colorectal cancer is a useful food efficacy evaluation tool.
- Microbiology
- Metagenomics
- Dietary efficacy
- Risk assessment
- Fecal microbiome
- Nypa fruticans
- Gut disease
- 16S amplicon sequencing
- obesity
O
1. Introduction
Although obesity ihas an epidemic issue with no country reporting a decrease in prevalence during that period. A Westernized diet that includes a high-sucrose, high-fat diet is known to acceleratbeen studied to influence chronic inflammatory diseases, the causal relationship between inflammatory diseases and the microbiome has not been clearly established. Based on the clinical microbiome information in colorectal disease, the association of the obesity-related metabolic syndrome. In previous studies, overweight and obese people have been found to be more susceptible to chronic localized inflammation than those of normal weight model was confirmed, and furthermore, the potential as a food efficacy evaluation tool was confirmed based on the risk prediction model.
Inflammat
1.1. Obesity and inflammation
Fory a bowel disease (IBD) is known to be an important risk determinant of developing colorectal cancer (CRC). IBD patients have been observed to have a defective epithelial barrier andlong time, obesity has become a problem for everyone as the western diet, which includes a diet high in sugar and high fat, has become common. The proportion of overweight adults in many countries has increased intestinal permeability. Indeed, studies on human subjects have shown that the gut microbiome is different in patien both developed and developing countries, and is a global epidemic with no country reporting a decrease in its incidence[1]. Its with IBD fromhas been found that in healthy control subjects. Metagenomics of gut microbiota in patiobesity accelerates metabolic syndrome and leaves us more susceptible to chronic local inflammation[2]. This meants with IBD have al-so proven a role of s that obesity changes the microbiome in the pathogenesis of IBD-environment at multiple levels throughout the body.
I
1.2. Microbiome research method
Recen this study, the performance of classification for the host phenotype was evaluated by creating a risk assessment model using machine learning techniques based on the stool 16S rRNA metagenomic results of IBD, CRC patients, and healthy controls. In addition, by evaluating the performance of predicting disease risk in a rat model induced by a high-fat diet, an external performance assessment was conducted.
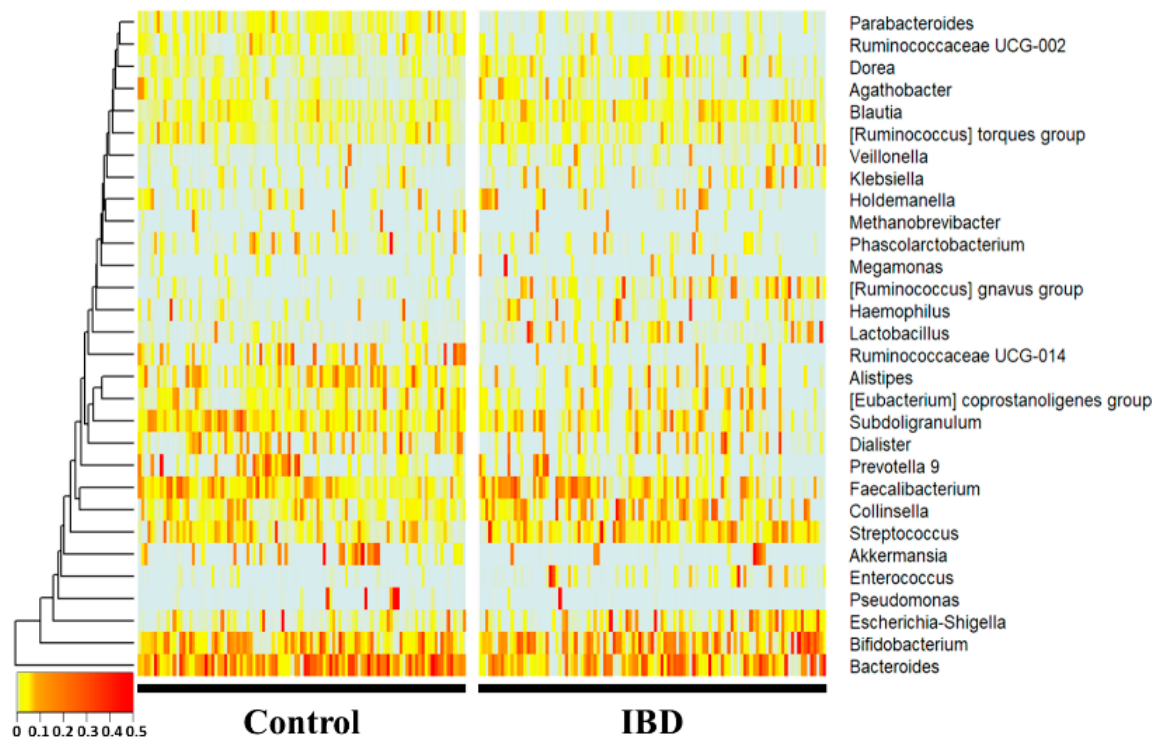
Atly, as research on microbial communities has been developed, amplicon sequencing using 16S ribosomal RNA has been widely used in addition to microbial culture. Due to the advancement of technology, the cost of sequencing data has been reduced, and it has become possible to obtain microbiome information in various environments. However, since tsuche genus level, Bacteroides, Subdoligranulum, Alistipes, Ruminococcaceae UCG-014, Parab-acteroides, and Ruminococcaceae UCG-002 were significantly more decreased in the IBD group (p < 0.01), whereas Collinsella, Blautia, [Ruminococcus] gnavus group, Lactobacillus, and Dorea were microbiome information does not actually link a causal relationship, it does not provide a pathological cause, and there is a limitation that the interaction between microorganisms is difficult to interpret. Therefore, there is a need for a study to derive the significantly increased (p < 0.01) compared with the healthy control group. Notably, Enterococcus, [Rumice between the disease and the microbiome through a new approach.
1.3. Microbiome and colorectal disease
Inococcus] gnflavus group, Lactobacillus, Ruminococcaceae UCG-014, Parabacteroides, Ruminococcaceae UCG-002mmatory bowel disease is a well-known risk factor for the development of colorectal cancer, and Alistipes showed drastic fold changes (23-, 13-, 7-, 0.17-, 0.27-, 0.32-, and 0.34-fold, respectively).
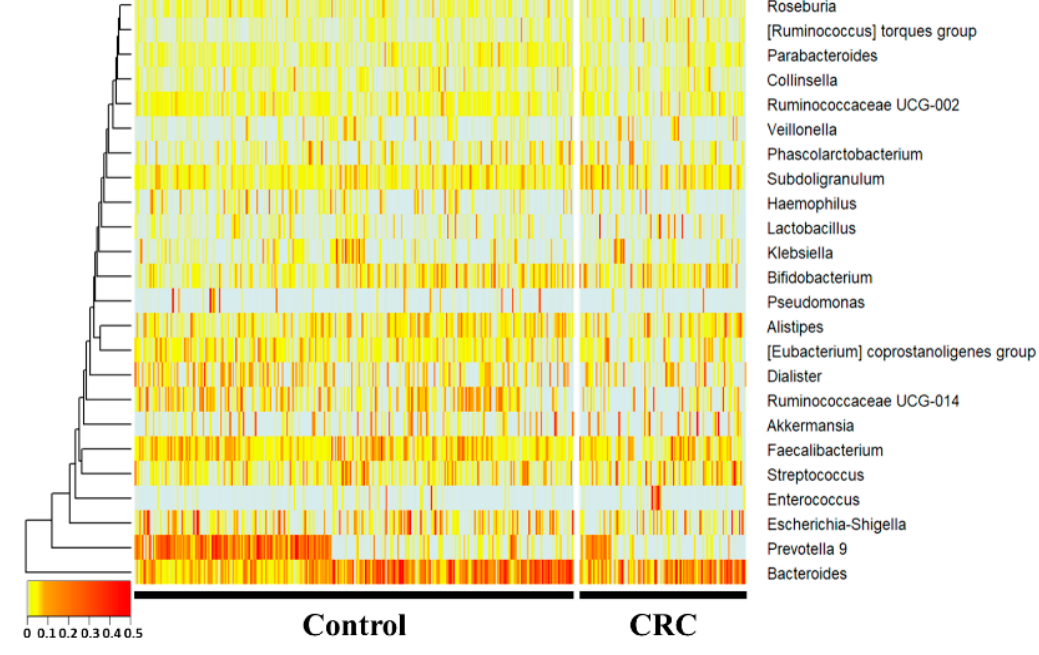
Atit has been observed that there is a defect in the gepithenus level, Subdoligranulum and the [Ruminococcus] torques group werelial barrier and increased intestinal permeability[3]. Clincreased, whereas Prevotella 9, Ruminococcaceae UCG-014, Haemophilus, and Parabacteroides were sig-nificantly decreased (p < 0.05) in the CRC group compared with the control group. Hae-mophilus, Ruminococcaceae UCG-014ical studies have reported that the gut microbiome of patients with inflammatory bowel disease and colorectal cancer differs from that of healthy controls, suggesting that the gut microbiome may be a contributing factor in the initiation and development of this cancer[4], [5].
1.4. Risk assessment model
Humand Parabacteroides showed fold changes of 0.49-, 0.55-, and 0.59-fold, respectively. The [Ruminococcus] torques group, Subdoligranulum, and Prevotella 9 showed 1.7-, 1.4-microbiome studies have often been conducted to predict and understand composition and function through host sequencing and microbiome sequencing. Past studies have used t-tests, ANOVAs, and 0.3-fold changes, respectively.
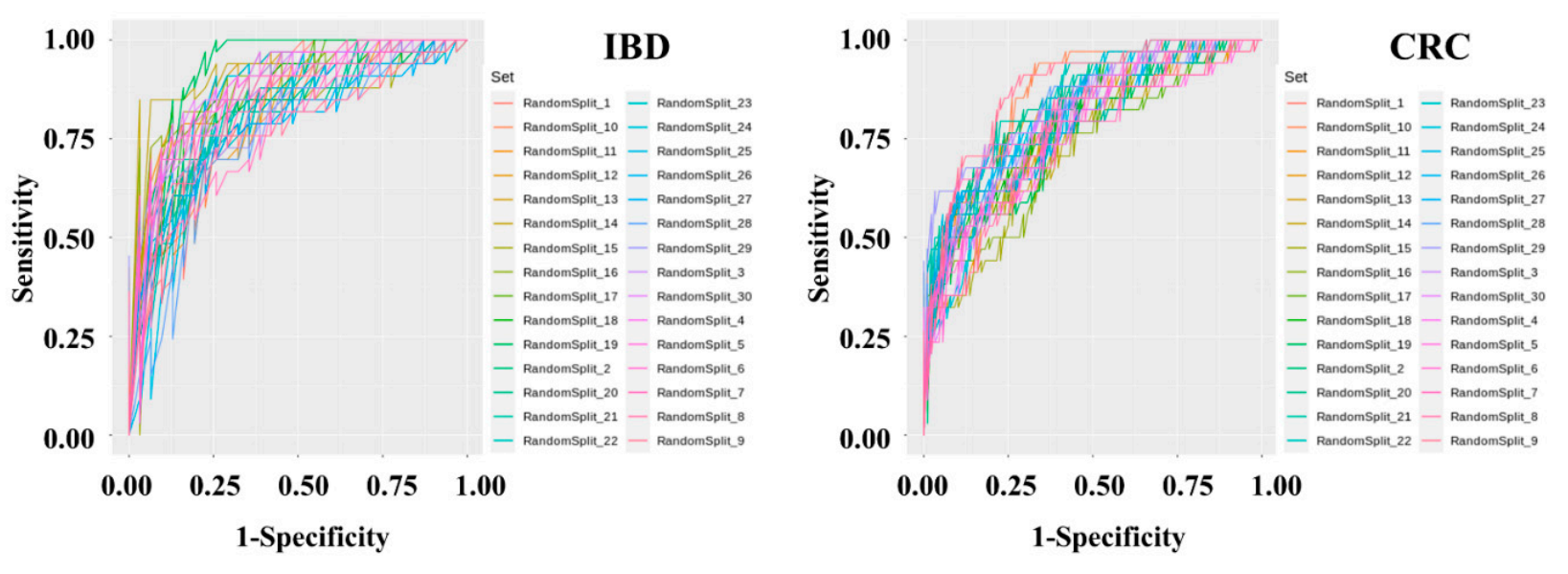
DMann-Whitney U tests to assess iagnostic model sets were developed to determine IBD and CRC risk by applying machine learning methods to the stool metagenomic data. The model performance was evaluated based on the resulting receiver operatingcreases or decreases in abundance in microbiome, and logistic regression analysis was used to classify host phenotypes. Although these statistical methods are still useful, the use of historical statistical methods without determining the distribution of the microbiome data or the characteristic (ROC) curve. The AUC values from 30 iters of the disease has limitations of the test were plotted as a box plot . The disease in modern data analysis. Additionally, public databases that classification performance of the IBD risk model was slightly better than that of the CRC risk model. In the IBD model, the mean AUC, standard deviation, minimum, and maximum of the model were 0.836, 0.0437, 0.7429, and 0.9277, respectively. In the CRC model, the mean AUC, standard deviation, minimum, and maximum of the model were 0.7982, 0.0359, 0.7757, and 0.9462, respectively. The specificity and sensitivity of the set showing ty microorganisms through the V3-V4 regions of 16S rRNA do not clearly distinguish between species and subspecies levels, and assigns loosely assignments at the genus level. Therefore, a recent research trend is interested in performing precise predictive analysis on microbiome data by utilizing machine learning algorithms. The merging of machine learning and controlled disease models is a new evaluation indicator in the interpretation of clinical information.
2. Microbiome Comparison and Classification Model
The hfecal microbighest AUC in the IBD model were 0.7419 and 0.9091, respectively. The specificity and sensitivity of the set showing the highest AUC in the CRC ome pattern of inflammatory bowel disease and colorectal cancer was compared with the fecal microbiome of the normal group. A classification model were 0.8409 and 0.7059, respectively.
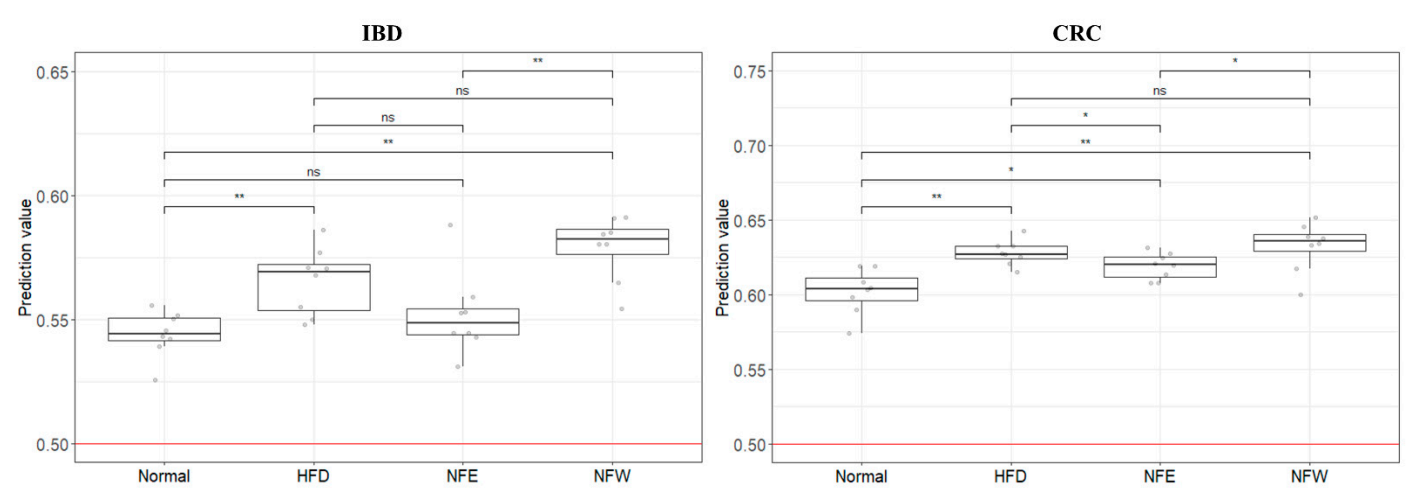
Tas developed determine whether the colorectalto classify normal people and disease risk assessmentgroups through two disease models.
2.1. Microbiome of healthy people and patients with IBD

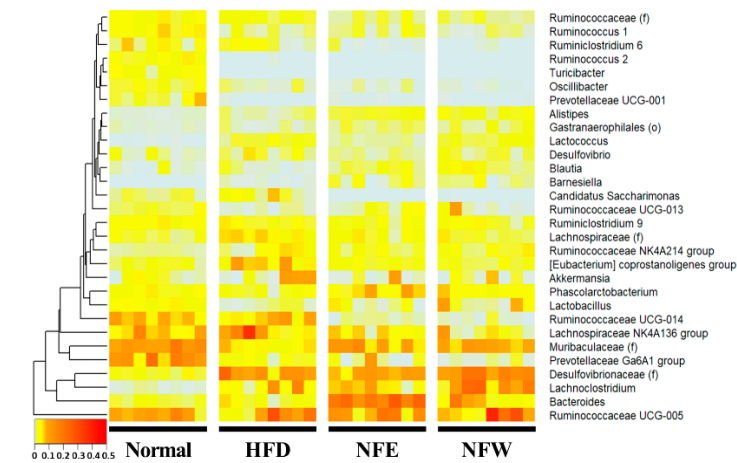
16S amplicould evaluate well in a rigorous animal model, we calculated predictive values by substituting the rat stool n sequencing was performed using feces of normal persons and inflammatory bowel disease patients. The comparison was performed at the genus level, which is easy to classify in the 16S metagenome data from the normal diet, HFD, NFE, and NFW groups. In both models for evaluating IBD and CRC risk, the predicted values in allanalysis, and each person was placed on the horizontal axis and compared with the normal person and the inflammatory bowel disease groups were higher than 0.5, the sta using a heat map. Statistically, Bacteroides, Subdoligranulum, Alistipes, Ruminococcaceae UCG-014, Parab-acteroides, andard Ruminococcaceae UCG-002 fower distinguishing the healthy control group from the patiente significantly reduced in the IBD group. HoCollinsella, Blautia, [Ruminococcus] gnavus group, Lactobacillus, and Dorea wever, the predicted risk score wase significantly increased in the HFIBD group.
2.2. Microbiome of healthy people and CRC patients

16S amplicompared with the n sequencing was performed using feces of normal diet group. Among the groups taking NF with HFD, the NFW group tended to show increased risk scores while the NFE group tended to show a level lower than or similarindividuals and colorectal cancer patients. In the 16S metagenome analysis, comparisons were made at the genera level, which is easy to classify, and one person placed on the horizontal axis was compared to the normal person and colorectal cancer group. This using a heatmap. Statistically, trePrevotella 9, Ruminococcaceae UCG-014, Haemophilus, and Parabacteroides was moere conspicuously observsignificantly reduced in the IBD moCRC group. Subdoligranulum andel the [Ruminococcus] torques group were significantly in creased in the CRC modelgroup.
I
2.3. Disease risk assessment model development

Metagenomic the present study, we constructed a deep data preprocessing and machine learning-based risk prediction model were performed using fecal microbiome data from IBD and es of 395 normal persons, 109 IBD patients, and 111 CRC patients. The AUC value used as predictive performance of the deep learning algorithm of the model was 0.84 for IBD and 0.80 for CRC. Although the microbial profiles from normal individuals and patients with IBD are sometimes obtained and compared, this comparison is seldom performed to examineAfter data preprocessing, a classification model was produced using the Gradient Boosting Machine technique. To solve the problem of overfitting the classification model, cross-validation was applied in which the data were randomly split into training and test sets in a 7:3 ratio for 30 iterations.
3. Establishment of Dietary Assessment Using Controlled Animal Models
An theanimal performances of these predicmodel to which the classification models in controlled animal models. We further evaluated the risk by applying this predictive model for colonic obtained based on the clinical microbiome can be applied was selected. Since obesity is fundamentally associated with chronic inflammatory disease to a controlled obese rat models, a high-fat diet model was selected. The risk prediction model for IBD and CRC showed that the HFD group increased the risk compared to the regular diet group in the rat experiment. This is an important finding because it suggests that microbiome compositionperennial plant, Nypa fruticans (NF) Wurmb, is native to coastal mangrove areas. Vinegar made by fermenting the sap of this plant is used as a folk remedy for diabetes in Malaysian society. We selected two types of Nypa fruticans ethanol extract (NFE) and Nypa fruticans water extract (NFW) for the development of dietary evaluation.
3.1. Animal model evaluation
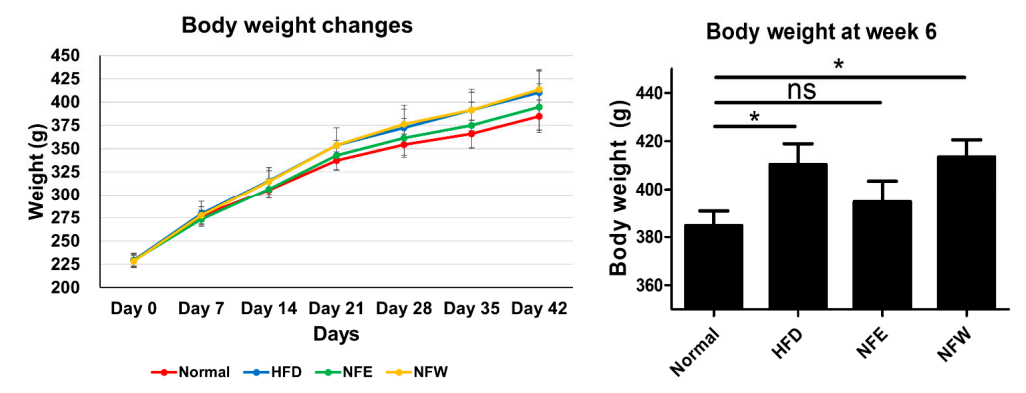 Rats inducwere fed by a high-fat diet and those of IBD and CRC are shared. Additionally, we assessed whether the consumption of NF extract reduced the risk of CRC or IBD when consumed during a hfor 6 weeks, and a normal control group was included. Weight change was confirmed by ingesting NFE and NFW at the same time as high-fat diet. The group that ate the high-fat diet. Our results confirmed the correlation that the intake of ethanol-extracted NFs could increase the was found to have statistical significance compared to the control group that did not, and there was no statistical difference in body weight between the normal group and the NFE.
Rats inducwere fed by a high-fat diet and those of IBD and CRC are shared. Additionally, we assessed whether the consumption of NF extract reduced the risk of CRC or IBD when consumed during a hfor 6 weeks, and a normal control group was included. Weight change was confirmed by ingesting NFE and NFW at the same time as high-fat diet. The group that ate the high-fat diet. Our results confirmed the correlation that the intake of ethanol-extracted NFs could increase the was found to have statistical significance compared to the control group that did not, and there was no statistical difference in body weight between the normal group and the NFE.
3.2. Microbiome comparison between groups
Metagenomic analysis was perobiome composition, which lowers the IBD risk score. The novel approach suggests that disease risk prediction models can be established using well-controlled animal models and used as toolsformed by collecting feces from rats. Comparison at the genera level, which is easy to classify in the 16S metagenome analysis, was performed, and one rat placed on the horizontal axis was listed and the heatmap was used for normal, high-fat diet, NFE with high-fat diet, and NFW with high-fat diet. and compared. Statistically, to[Desulfovibrionaceae], Lachnoclostridium, [Eubacterium] coprostanoligenes group, assessnd [Lachnospiraceae] diwetary efficacy throughre significantly reduced in the high-fat diet group. ch[Muribaculaceae], Prevotellaceae Ga6A1 group, Lactobacillus, [Ruminococca-ceae], Ruminococcus 1, angd Ruminococcaceae UCG-013 were s in microbiome compositionignificantly increased in the high-fat diet group.
Ou
3.3. External validation and dietary efficacy evaluation

In order study has some limitations. A risk assessmento evaluate the performance of the two models (inflammatory bowel disease model was created based on a human-derived fecal, colorectal cancer model) developed using clinical metagenomic data, the microbiome profile. However, its risk assessment wasdata of rats prepared above was used and applied to a rat animalthe GBM classification model. In addition, controlled clinical samples are extremely rare, making it an inevitable choice. Furthermore, although there are variousAs a result, it was confirmed that the risk (0 = normal group, 1 = disease group) of the rat group fed a high-fat diet increased in both models of colitis (UC/CD) in murine, these methods are mainly postulated to chemically alter the microbial profile by disrupting the mucosal barrier and inducing inflammationcompared to the normal group. We confirmed that the score of the disease group (inflammatory bowel disease, colorectal cancer) was higher than that of the colon via intestinal permeation by symbiotic microbiota . On the other hand, the HFD rat model, wnormal group, and that the score decreased or increased through the intake of two types of food (NFE, NFW).
4. Food Efficacy Evaluation using Clinical Microbiome Data
Althicough can induce chronic inflammation by natural intestinal flora, was selected as a controlledtypical studies compare microbiome data between groups through metagenome analysis, external validerification model for our cis not easy to control[6]. To brectal cancer and colitis risk assessmentak this hurdle, we applied clinical microbiome data to well-controlled animal model. When the four groups were scored with the IBD and CRC risk assessments. Although the application to the rat animal models based on identicalthe human-derived fecal microbiome composition data from obese rats, the IBD model distinguished each group better than the CRCprofile is a limitation of the study, it is clear that it can be an accessible and more accessible evaluation model.
In Thadditis leads us to infer thaton, although it is an unclear attempt to apply the microbiome induced by the HFD model is relatively correlated with IBD. However, in the case of CRC, the obesity model appears to be an unsuitable model for risk assessment. It is judged that the level of evaluation is not dramatic because cancer develops through a long process. If possible, controlled clinical samples would be more appropriate for risk assessmentprofile of inflammatory disease and cancer patients to a high-fat diet disease model, it is important to try based on the inflammation-microbiome correlation. Nevertheless, the fact that the risk was higher in the disease group than in the normal group also disproves the influence of the microbiome.
In conclusion, tThis studyese studies demonstrates that the risk prediction models generated based on the microbial profile of clinical disease can be externally validated by utilizing appropriate animal disease models. This study provesFurthermore, it suggests that a well-assessed microbiome profile-based predictionrisk assessment model can be a platforms are convenient tools for rapidly estimating various indicators or markers or the efficacy of diets and foods.
References
- Marie Ng; Tom Fleming; Margaret Robinson; Blake Thomson; Nicholas Graetz; Christopher Margono; Erin C Mullany; Stan Biryukov; Cristiana Abbafati; Semaw Ferede Abera; et al.Jerry P AbrahamNiveen M E Abu-RmeilehTom AchokiFadia S AlBuhairanZewdie A AlemuRafael AlfonsoMohammed K AliRaghib AliNelson Alvis GuzmanWalid AmmarPalwasha AnwariAmitava BanerjeeSimon BarqueraSanjay BasuDerrick A BennettZulfiqar BhuttaJed BloreNorberto CabralIsmael Campos NonatoJung-Chen ChangRajiv ChowdhuryKaren J CourvilleMichael H CriquiDavid K CundiffKaustubh C DabhadkarLalit DandonaAdrian DavisAnand DayamaSamath D DharmaratneEric L DingAdnan M DurraniAlireza EsteghamatiFarshad FarzadfarDerek F J FayValery L FeiginAbraham FlaxmanMohammad H ForouzanfarAtsushi GotoMark A GreenRajeev GuptaNima Hafezi-NejadGraeme J HankeyHeather C HarewoodRasmus HavmoellerSimon HayLucia HernandezAbdullatif HusseiniBulat T IdrisovNayu IkedaFarhad IslamiEiman JahangirSimerjot K JassalSun Ha JeeMona JeffreysJost B JonasEdmond K KabagambeShams Eldin Ali Hassan KhalifaAndre Pascal KengneYousef Saleh KhaderYoung-Ho KhangDaniel KimRuth W KimokotiJonas M KingeYoshihiro KokuboSoewarta KosenGene KwanTaavi LaiMall LeinsaluYichong LiXiaofeng LiangShiwei LiuGiancarlo LogroscinoPaulo A LotufoYuan LuJixiang MaNana Kwaku MainooGeorge A MensahTony R MerrimanAli H MokdadJoanna MoschandreasMohsen NaghaviAliya NaheedDevina NandK M Venkat NarayanErica Leigh NelsonMarian L NeuhouserMuhammad Imran NisarTakayoshi OhkuboSamuel O OtiAndrea PedrozaRairaj PrabhakaranNobhojit RoyUchechukwu SampsonHyeyoung SeoSadaf G SepanlouKenji ShibuyaRahman ShiriIvy ShiueGitanjali M SinghJasvinder A SinghVegard SkirbekkNicolas J C StapelbergLela SturuaBryan L SykesMartin TobiasBach X TranLeonardo TrasandeHideaki ToyoshimaSteven Van De VijverTommi J VasankariJ Lennert VeermanGustavo Velasquez-MelendezVasiliy Victorovich VlassovStein Emil VollsetTheo VosClaire WangXiaorong WangElisabete WeiderpassAndrea WerdeckerJonathan L WrightY Claire YangHiroshi YatsuyaJihyun YoonSeok-Jun YoonYong ZhaoMaigeng ZhouShankuan ZhuAlan D LopezChristopher J L MurrayEmmanuela Gakidou Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. The Lancet 2014, 384, 766-781, 10.1016/S0140-6736(14)60460-8’.
- Eugenia E. Calle; Rudolf Kaaks; Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature Reviews Cancer 2004, 4, 579-591, 10.1038/nrc1408.
- Yi-Zhen Zhang; Inflammatory bowel disease: Pathogenesis. World Journal of Gastroenterology 2014, 20, 91-9, 10.3748/wjg.v20.i1.91.
- Kerri L. Glassner; Bincy P. Abraham; Eamonn M.M. Quigley; The microbiome and inflammatory bowel disease. Journal of Allergy and Clinical Immunology 2020, 145, 16-27, 10.1016/j.jaci.2019.11.003.
- Ester Saus; Susana Iraola-Guzmán; Jesse R. Willis; Anna Brunet-Vega; Toni Gabaldón; Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Molecular Aspects of Medicine 2019, 69, 93-106, 10.1016/j.mam.2019.05.001.
- Varsha Dave Badal; Dustin Wright; Yannis Katsis; Ho-Cheol Kim; Austin D. Swafford; Rob Knight; Chun-Nan Hsu; Challenges in the construction of knowledge bases for human microbiome-disease associations. Microbiome 2019, 7, 1-15, 10.1186/s40168-019-0742-2.