Time-lapse microscopy imaging is a key approach for an increasing number of biological and biomedical studies to observe the dynamic behavior of cells over time which helps quantify important data, such as the number of cells and their sizes, shapes, and dynamic interactions across time. Label-free imaging is an essential strategy for such studies as it ensures that native cell behavior remains uninfluenced by the recording process. Computer vision and machine/deep learning approaches have made significant progress in this area.
1. Introduction
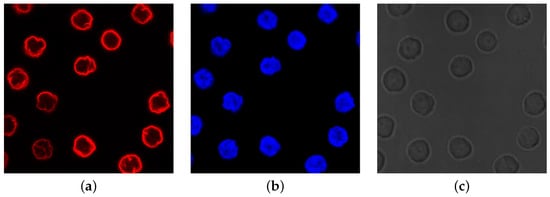
Microscopy is a fundamental research pillar enabling scientists to discover the structures and dynamics of cells and subcellular components. Most of these components are phase objects, which means they are transparent and colorless and cannot be visualized under a light microscope. To overcome this limit, a solution consists in staining the components with dyes, also known as fluorophores or fluorochromes. Such molecules absorb short-wavelength light, generally UV, and emit fluorescence light at a longer wavelength. This mechanism represents the basic premise of fluorescence microscopy techniques, and the fluorescence images show the specimen bright on a dark background. The staining techniques use different dyes to point out a specific cellular component. Then, the same specimen can appear with a red, blue, or green color and appearance, as depicted in Figure 1a,b.
As molecular genetic methodologies and tools advance, fluorescence techniques become more specific and can be applied to a larger set of different model organisms [
1,
2]. However, to ensure high-quality images, they require laborious and expensive sample preparation. Furthermore, the dyes reduce their luminance over time due to a light-induced degradation process called
photobleaching [
3]. Finally, fluorescent techniques are invasive and interfere with biological processes causing phototoxicity.
On the contrary,
Label-free Imaging (LI) techniques can visualize many cellular structures simultaneously with minimal sample preparation, phototoxicity, and no photobleaching, making them particularly suitable for live-cell imaging. Thus, LI provides measurements complementary to fluorescence imaging for several biological studies. Based on optical principles, these techniques measure the light phase change (i.e., the refractive index) passing through the specimen and converting it into intensity modulations producing
qualitative phase contrast images, as exemplified in
Figure 1c (the phase change information contained in LI images is non-linearly coupled with its luminance intensity and cannot be retrieved quantitatively. An image produced by LI techniques is a map of path-length shifts associated with the specimen, containing information about both the thickness and refractive index of its structure; for further details and explanations, see [
4]). Among the traditional techniques to phase change imaging, there are
Phase Contrast (PhC) [
5] and
Differential Image Contrast (DIC) [
6] based on the phase gradient method and differential interference contrast, respectively, to measure the refractive index. Another LI technique very similar to DIC is the
Hoffman Modulation Contrast (HMC) [
7]. Due to intrinsic limitations of the numerical conversion methods, phase contrast images contain artifacts [
8] (i.e., bright halo surrounding cell contours) and the “shade-off effect”, which produces low contrast inside the cells with an intensity very similar to the background. Although several methods have been developed to overcome these artifacts, automatic image processing of label-free images is still challenging, especially the segmentation task for separating cells from the background.
Figure 1. Example of differences in appearance of fluorescence and phase contrast microscopy images (culture of human lymphocyte cells [
9]): (
a) fluorescence image of nuclear envelope; (
b) fluorescence image of interior nuclei (DNA); (
c) phase contrast image of whole cells.
Unlike the previous techniques, quantitative LI provides higher contrast and reduces artifacts. Among them,
Quantitative Phase Imaging (QPI) techniques refer to the microscopes showing phase quantitative information. Details concerning QPI techniques can be found in [
10].
A further alternative to noninvasive techniques is Bright-Field (BF) microscopy. It represents the most straightforward configuration for the light microscope, which is not only cheaper but also does not require sample preparation [
11]. BF images provide information about the cellular organization, and they are preferred to visualize specimens with low contrast from the background (unlike in fluorescence) or with low resolution and magnification visualization of thin cellular components (unlike in phase contrast). Several studies describe the use of the BF channel in cell detection and automated image analysis of cell populations [
12,
13].
With the aim of observing the dynamic behavior of living cells over time, LI microscopes are also equipped with a real-time imaging tool named
time-lapse. Broadly speaking, time-lapse is a speed-up technique to observe events changing over time. This is usually realized by taking images at regular time intervals and merging them into a video. One of the most significant applications of time-lapse microscopy is cell population monitoring to study single-cell behavior in response to physiological or external stimuli and understand the underlying mechanisms. For example, in drug discovery and cancer research [
14], time-lapse microscopy is used to look at cell response to anti-mitotic drugs in terms of cell division and cell death. To achieve this goal, quantitative information on cell behavior needs to be obtained and analyzed [
15]. Cell proliferation, lineage, and fate are of primary importance among various cellular events. Image analysis of these biological processes is usually performed manually with suitable protocols [
16]. As manual analysis of a large volume of light microscopy images is slow, tedious, time-consuming, and subject to observer subjectivity, biological studies see an increased demand for reliable automatic imaging tools. As in many scientific disciplines, Artificial Intelligence (AI) has been changing how imaging data are processed and analyzed and how experiments are carried out. AI refers to artificial systems aiming to adapt previous knowledge to new situations and recognize meaning in data patterns. Machine Learning (ML) [
17] is a subset of AI methods that extracts valuable features from large data sets to make predictions or decisions on unseen data. An ML algorithm is not designed to solve a specific problem but rather to train a computer to solve problems. The training is data-driven. Deep Learning (DL) is a set of ML algorithms using a multi-layer “neural networks” to progressively extract higher-level features from data (the number of layers is the depth of the model, hence the terminology “deep learning”; for a quick overview on DL key concepts for microscopy image data, the reader can refer to [
18]).
2. Cell Segmentation
Image segmentation is the main task for producing numerical data from live-cell imaging experiments, thus providing direct insight into the living system from the quantitative cell information [
23,
24]. Cell segmentation is the process of splitting a microscopy image into “segments”, i.e., Regions Of Interest (ROIs), and produces an image where cells are separated from the image background by cell contours or cell labels. Accurate cell segmentations are crucial for many challenges involved in cellular analysis, including but not limited to cell tracking [
25], cellular features quantification, proliferation, morphology, migration, interactions, and counting [
26,
27,
28,
29].
Several steps are considered in the literature for achieving cell segmentation. Some authors [
8,
20,
30] consider crucial to initially perform an image reconstruction step, which produces images with higher contrast between foreground and background, increasing the success of the subsequent image processing tasks. The image formation model in phase contrast microscopy can be studied to reduce artefacts and solve the inverse problem through a regularization approach [
8]. Some ML-based methods are reviewed by Vicar et al. [
20] for PhC and DIC images, while De Haan et al. [
30] present an overview of how DL-based frameworks solve these inverse problems in optical microscopy.
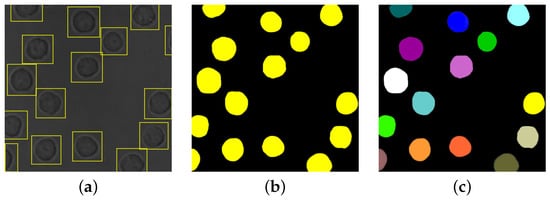
Cell detection (or identification) is also frequently adopted previous to segmentation [
20,
22], with the aim of locating the cells in the image (e.g., via bounding boxes, as exemplified in
Figure 2a).
Figure 2. Results of different image processing tasks: (a) cell detection; (b) cell semantic segmentation; (c) cell instance segmentation.
Some specific types of segmentation are frequently considered for cellular images [
20,
31,
32,
33]. Semantic segmentation identifies the object (i.e., the cell) category of each pixel for every known object within an image, as exemplified in
Figure 2b. Instance segmentation, instead, identifies the object instance (i.e., the cell with specific features) of each pixel for every known object within an image [
34]; an example is given in
Figure 2c. It should be observed that the problem of cell instance segmentation is sometimes intended as joint cell detection and segmentation (e.g., see [
32]), while other times (e.g., see [
20]) instant segmentation is used as a synonym for single cell segmentation.
3. Event Detection and Classification
Even though segmentation remains the core of the subsequent imaging tasks, automated analysis of microscopy image sequences often bypasses the segmentation task. Usually, the detection of cell events under investigation is performed directly from heuristically generated ROIs. Detecting changes in cellular behavior plays a central role in different studies, where the focus is on identifying the changes in cellular growth, mitosis, and death. Such changes may be related to cell shape, division, and movement. They cannot be detected in a single image but require the analysis of video or time-lapse sequences. The difficulty primarily relies on the wide spatial-temporal variability of such phenomena, which requires suitable methods to handle time-varying phenomena. Nevertheless, the infinite spectrum of possible events faces an inherent shortage of labeled data.
Automatic and robust approaches to detecting the time and location of cell events from image sequences often make use of the classification task. In microscopy, classification refers to identifying and distinguishing different cell types or states. Classification between other cells, types of tumors (benign or malignant), types of cell states (mitosis detection, alive-dead classification), and types of CIC (Cell-In-Cell) structures [
47] are some typical applications.
4. Cell Tracking
Object tracking consists in locating and monitoring one or more objects of interest and their behavior over time [
21]. The image sequence containing cells can be acquired at specific time intervals using the time-lapse technique. When discussing cell tracking, it is generally assumed that segmentation or detection and classification have been performed.
5. Software
In Table 1, we provide links to existing publicly available software.
6. Data
In Tables 2, 3, and 4, we provide details of available annotated data for cell segmentation, event detection, and tracking in traditional label-free images.