Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Cristian Arriaga-Canon.
SARS-CoV-2 is a coronavirus family member that appeared in China in December 2019 and caused the disease called COVID-19, which was declared a pandemic in 2020 by the World Health Organization. Concerning nucleic acid-based therapy, RNA therapy has shown promising results in treating several human malignances, including viral diseases. There are different approaches to targeting RNA in the human body: small interfering RNAs (siRNAs), microRNAs (miRNAs), antisense oligonucleotides (ASOs), and CRISPR/Cas systems, among others.
- SARS-CoV-2
- transcriptomics
- RNA therapeutics
- precision medicine
1. Small Interfering RNAs
These noncoding double-stranded transcripts are ~20 nucleotides long and mediate the specific suppression of transcripts [138,139][1][2] by binding to the RNA-induced silencing complex (RISC) [140][3]. The single-stranded siRNA guides RISC to its target RNA, leading to gene silencing [141][4]. In the pandemic caused by SARS-CoV in 2003, siRNAs effectively reduced viral replication in infected cells [142,143,144][5][6][7] and in vivo models [145][8]. Regarding SARS-CoV-2, in a study in which primary human tracheal cells (hpTCs) expressing the S glycoprotein were treated with siRNAs [146][9], a significant reduction in this protein was reported, cell viability and proliferation remained unchanged, and modified siRNAs with cholesterol moieties were used for cells to directly uptake siRNA from the medium, a useful approach for future therapeutic applications in which some delivery methodologies may cause toxic effects [107,147][10][11]. In the search for lower-toxicity delivery systems, a lipid nanoparticle was used to deliver siRNAs targeting RNA-dependent RNA polymerase (RdRp), helicase (Hel), and the 5′ untranslated region (5′-UTR) in the SARS-CoV-2 genome using a K18-hACE2 mouse model [148][12], and a 90% reduction in viral replication was observed. COVID-19-associated symptoms were lower in treated mice, which also exhibited higher overall survival, demonstrating that siRNAs are a useful therapy against SARS-CoV-2. In this study, the siRNA delivery was made through a formulation described previously [149][13] in which the polyethylene glycol (PEG)2000–C16 ceramide conjugate was prepared using the hydration of freeze-dried matrix (HFDM) method to form liposomes; furthermore, intravenous delivery of this conjugate proved successful delivery of siRNA in the lung epithelium and achieved gene silencing in these cells [150][14]. Nevertheless, the precise transfection mechanism into the cells remains to be described. Using another in vivo model, an siRNA targeting the RdRp gene that significantly reduced viral replication and lung inflammation in Syrian hamsters was reported [151][15]. This siRNA administration was performed by inhalation, a tool that could be a promising way to deliver drugs for respiratory diseases; in this case, a cationic dendrimeric peptide (KK-46) proved to be successful in the intracellular delivery of siRNA with very low toxicity. Therapeutic siRNAs have a confirmed efficacy against SARS-CoV-2; nevertheless, clinical studies are needed to support siRNA therapy as safe and efficient for COVID-19 in humans.
2. MicroRNAs (miRNAs)
In contrast to siRNAs, miRNAs are single-stranded RNAs with a length of 19–25 nucleotides [152][16] that can regulate not only one target, but may have different targets [153,154][17][18]; nevertheless, both noncoding RNAs share a similar mechanism of action through their association with RISC and Dicer for post-transcriptional inhibition of mRNA [155,156][19][20]. Such miRNAs have shown to play key roles during human diseases such as cancer [157,158][21][22] and in infectious diseases [159,160][23][24]. In SARS-CoV-2 infection, expression of more than 70 miRNAs were deregulated in peripheral blood [161][25], suggesting the importance of these noncoding RNAs in the infectious process. They have also been proposed as potential biomarkers of the severity of COVID-19 [162,163][26][27], as well as for therapeutic approaches to SARS-CoV-2 infection [164,165][28][29].
3. Antisense Oligonucleotides
ASO-based therapy has been successfully used for human viral diseases [166,167][30][31]; in general, ASOs are single-stranded DNA sequences of ~20 nucleotides in length that bind to complementary RNA via Watson–Crick base pairing [168,169,170][32][33][34]. ASOs can modify RNA functions through mechanisms that depend on their chemical structure and modifications [170,171][34][35]. ASOs can interfere with mRNA translation [172,173][36][37] and splicing [174][38] but can also recruit RNase H1 and H2 to induce RNA degradation [81,175][39][40]. Several ASO-based therapies depend on these mechanisms to maintain low levels of the target sequence [176,177,178][41][42][43]. Therapies against human viruses such as hepatitis B [179[44][45],180], Ebola [181][46], influenza A [182][47], and West Nile virus [183][48], among others, are being developed and have demonstrated high specificity and efficacy, suggesting a promising landscape for ASO-based therapies.
Regarding SARS-CoV-2, in a study in which ASOs were designed using in silico tools to disrupt the interactions between SARS-CoV-2 5′-RNAs and host proteins needed to regulate the viral infection cycle [184][49], in vitro experiments on the human cell line Huh7.5.1 reported an ~50% decrease in the SARS-CoV-2 RNA yield in treated cells. They also designed ASOs that targeted structurally conserved regions of SARS-CoV-2 RNA (ORF1 and N) that reduced the viral infection ratio by 50% in the human cell line Caco-2, which was derived from human colon carcinoma. Cells were transfected with ASOs using lipofectamine; the transfection mechanism was the formation of cationic liposomes that enabled the internalization of anionic ASOs. Then, in contact with the cell membrane, liposomes created endosomes to enter the cell [185][50]. The innovative in silico tool they proposed for ASO design succeeded in targeting different regions of the viral genome and reduced the viral yield and infection.
Peptide nucleic acids (PNAs) are another type of ASO that consist of a polypeptide backbone with nucleic acid bases attached as side chains and are used as DNA/RNA analogs to inhibit transcription and translation [186][51]. In a study conducted by Ahn and collaborators [187][52] using programmed −1 ribosomal frameshifting (−1 PRF) in the SARS-CoV genome, a region controlling the synthesis of viral replicase proteins [188][53] was targeted using PNAs in HEK293 cells; inhibition of viral replication was observed compared to the control, suggesting that -1 PRF is an important target for SARS-CoV treatment. Cells were transfected using FuGENE, a cationic polymer that creates complexes with anionic ASOs. The complexes could create endosomes in the cell membrane to penetrate the cell [185,189][50][54]. Even though ASOs have been demonstrated to be efficient and specific in targeting SARS-CoV-2 in vitro and in vivo, further experiments are needed to confirm their potential use as therapeutic agents in humans against SARS-CoV-2.
4. CRISPR-Cas
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) systems are an adaptive immune system in prokaryotes [190,191,192][55][56][57] that was first described in the late 1980s and early 1990s [193][58]. Currently, the therapeutic gene-editing applications of this technology are a promising tool to treat several human malignances [194,195,196][59][60][61] and viral diseases [197,198][62][63]. Among CRISPR-Cas effectors, two recently discovered effectors are specific to targeting single-stranded RNA (ssRNA), type VI-A (Cas13a) and type VI-B (Cas13b) [199][64], which were the first discovered among CRISPR-Cas systems that specifically targeted RNA [175,200,201][40][65][66]. One of the RNA viruses previously targeted by CRISPR-Cas13 is HIV-1. Yin and collaborators [119][67] observed a strong inhibition of HIV infection in human HEK293T cells, reduced viral RNA and protein levels, and reduced viral DNA integration after infection with mature HIV-1 viral particles compared to control treatment.
Regarding SARS-CoV-2, Abbott and collaborators [202][68] reported a CRISPR-Cas13-based strategy called prophylactic antiviral CRISPR in human cells (PAC-MAN), which revealed a group of 6 crRNAs able to target 91% of all sequenced coronaviruses and a group of 22 crRNAs that could target 100% of the sequenced coronaviruses. In experiments on the human lung epithelial A549 cell line using reporters of synthetic fragments of SARS-CoV-2 RdRP or nucleocapsid (N) genes fused to GFP, pools of four of the predicted crRNAs were used to target these constructs, resulting in a 70% reduction in the reporter gene expression. The use of a pool of crRNAs could be helpful in the case of a live infection in which mutation or the presence of viral variants could escape from a single crRNA. In contrast to this virus-free model, in a study conducted by Fareh and collaborators [203][69], CRISPR-Cas13b was used to target the mRNA encoding the N protein in replication-competent particles of SARS-CoV-2 using the Calu-3 cell line derived from human lung adenocarcinoma. After viral infection, the silencing efficiency, as measured by a tissue culture infectious dose that inhibited 50% of the virus growth in an infectivity assay (TCID50), showed that replication was significantly lower compared to the control, even at the longest time measured (48 h). Similar results were observed in Vero cells, a nonhuman mammalian cell line in which pools of four different crRNAs were used to target several regions of the SARS-CoV-2 genome; 80–90% viral suppression was achieved with all tested crRNAs, as well as suppression of the SARS-CoV-2 variant D614G and B.1.1.7, known as the UK variant. In this study, the cell line expressing Cas13d was transduced by lentiviral infection with the pooled crRNAs. The lentiviral transduction mechanism was based on the viral infection of the cells by the virion; the virion could enter the cell by endocytosis or by binding to specific receptors, then the nucleic acid contained inside the virion was released inside the cell [204,205][70][71].
In another study using a replication-competent SARS-CoV-2 virus [206][72], CRISPR-Cas13a crRNAs were designed to target RdRp and N genes in the Vero E6 cell line. A combination of two crRNAs targeting the N gene reduced 72% of cell death in infected cells, while the mRNA of RdRp and N genes exhibited a reduction of 93.7% and 94.1% in copy number, respectively. After successful in vitro assays, in vivo experiments were performed on Syrian hamsters using a crRNA targeting the N gene. Administration of mRNA encoding Cas13a and crRNA formulated with poly-beta amino ester, which was suitable for delivery through inhalation, was performed using a cone nebulizer; 20 h later, hamsters were infected with SARS-CoV-2. Six days later, a 57% decrease in the viral N mRNA copy number was observed compared to the control, suggesting CRISPR-Cas13a as a potential antiviral therapy against SARS-CoV-2. The in vivo delivery of mRNA-encoding CRISPR-Cas13a and crRNAs was performed using hyperbranched poly (beta amino esters) (hPBAEs); these compounds create polyplexes with the anionic RNAs that allow endosome formation in the cell membrane to penetrate the cell [207][73].
CRISPR-Cas13 systems have been demonstrated to be efficient for SARS-CoV-2 RNA knockdown not only in in vitro virus-free models, but also when using replication-competent viral particles in human cell lines and in vivo models (Figure 31). The approach to using CRISPR-Cas13 goes beyond therapy against COVID-19; due to its specificity and flexible design against different genomic targets, it could be used to treat different human viral diseases and represents a powerful weapon against future pandemics.
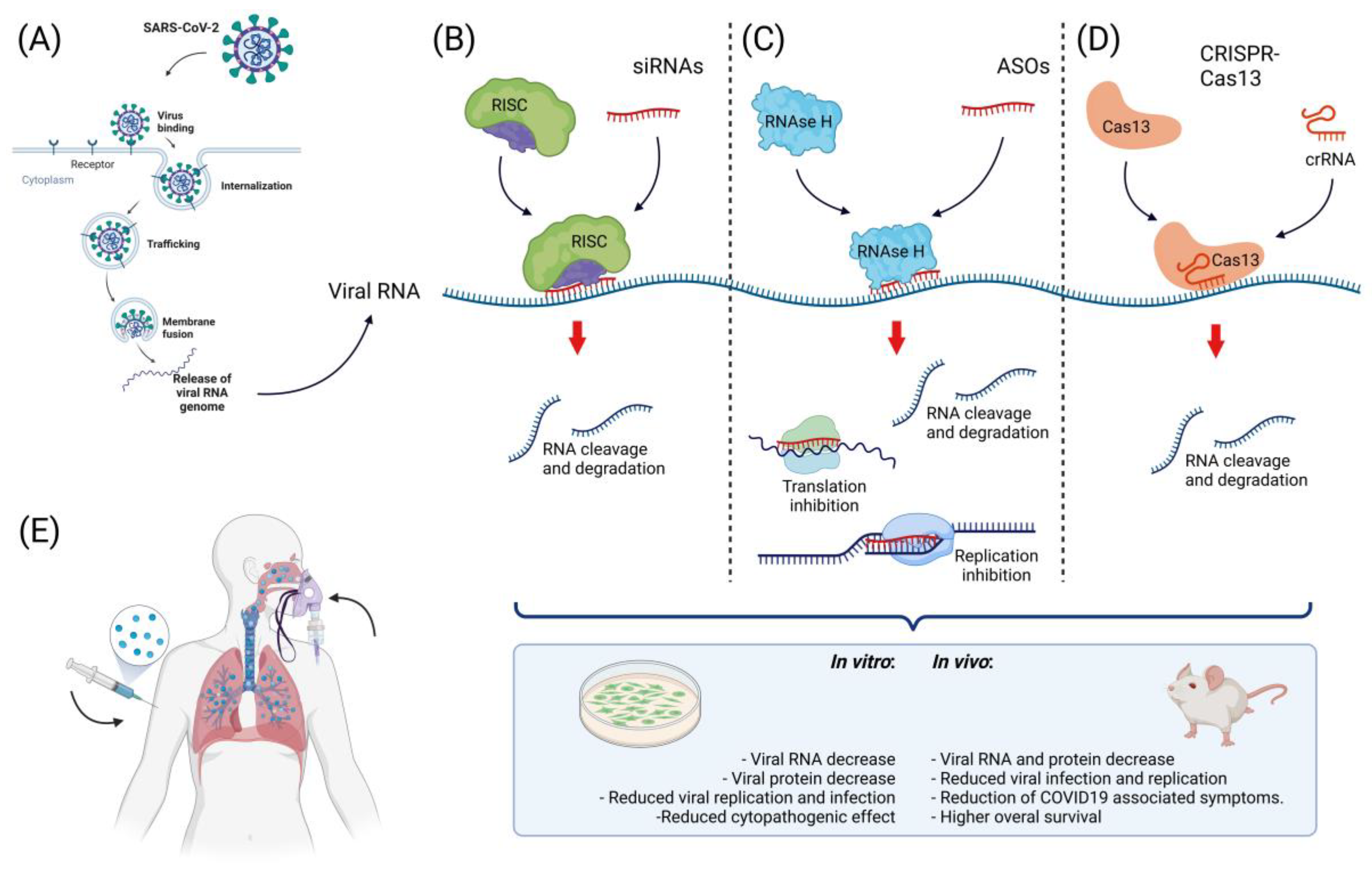
Figure 31. Approaches to nucleic acid-based therapy against SARS-CoV-2. (A) Scheme of viral infection with SARS-CoV-2. After viral binding to cell receptors, the viral particle is internalized and then releases viral RNA to the cytoplasm, where viral components are synthesized. At this point, free viral RNA in the cytoplasm can be targeted. (B) One of the strategies for targeting viral RNA inside the cell is the use of siRNAs; these small molecules (~20 nt) regulate RNA degradation through their interaction with the RISC complex. The activated RISC complex binds to its target, resulting in RNA cleavage and degradation; the use of siRNAs targeting SARS-CoV-2 has been successful in vitro and in vivo. (C) Another approach to targeting this virus is the use of ASOs; these molecules mediate RNA cleavage by recruiting RNAse H and interfere with replication, transcription, and transduction, resulting in decreased viral RNA and protein levels. ASOs have been successfully used to target SARS-CoV-2 in vitro. (D) The use of genome-editing tools has increased the possibilities for SARS-CoV-2 treatment, while the use of CRISPR-Cas13 has been successful in targeting this virus in vitro and in vivo, resulting in a reduction in viral RNA and protein, a decrease in viral infection, and a decrease in COVID-19-associated symptoms. (E) For the successful targeting of viral RNA, the use of an efficient delivery method that guarantee the proper release of the therapeutic nucleic acid to the cell is important. For the internalization of negatively charged nucleic acids into the cell, the use of cationic compounds and lipid-based delivery methods have been successfully used for this particular therapy via intravenous or inhalation delivery.
References
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411, 494–498.
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA Interference Is Mediated by 21- and 22-Nucleotide RNAs. Genes Dev. 2001, 15, 188–200.
- Ameres, S.L.; Martinez, J.; Schroeder, R. Molecular Basis for Target RNA Recognition and Cleavage by Human RISC. Cell 2007, 130, 101–112.
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51.
- Shi, Y.; Yang, D.H.; Xiong, J.; Jia, J.; Huang, B.; Jin, Y.X. Inhibition of Genes Expression of SARS Coronavirus by Synthetic Small Interfering RNAs. Cell Res. 2005, 15, 193–200.
- Wu, C.-J.; Huang, H.-W.; Liu, C.-Y.; Hong, C.-F.; Chan, Y.-L. Inhibition of SARS-CoV Replication by SiRNA. Antivir. Res. 2005, 65, 45–48.
- Wang, Z.; Ren, L.; Zhao, X.; Hung, T.; Meng, A.; Wang, J.; Chen, Y.-G. Inhibition of Severe Acute Respiratory Syndrome Virus Replication by Small Interfering RNAs in Mammalian Cells. J. Virol. 2004, 78, 7523–7527.
- Li, B.; Tang, Q.; Cheng, D.; Qin, C.; Xie, F.Y.; Wei, Q.; Xu, J.; Liu, Y.; Zheng, B.; Woodle, M.C.; et al. Using SiRNA in Prophylactic and Therapeutic Regimens against SARS Coronavirus in Rhesus Macaque. Nat. Med. 2005, 11, 944–951.
- Gallicano, G.I.; Casey, J.L.; Fu, J.; Mahapatra, S. Molecular Targeting of Vulnerable RNA Sequences in SARS CoV-2: Identifying Clinical Feasibility. Gene Ther. 2020, 29, 304–311.
- Dowdy, S.F. Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol. 2017, 35, 8.
- Juliano, R.L. The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548.
- Idris, A.; Davis, A.; Supramaniam, A.; Acharya, D.; Kelly, G.; Tayyar, Y.; West, N.; Zhang, P.; McMillan, C.L.D.; Soemardy, C.; et al. A SARS-CoV-2 Targeted SiRNA-Nanoparticle Therapy for COVID-19. Mol. Ther. 2021, 29, 2219–2226.
- Wu, S.Y.; Putral, L.N.; Liang, M.; Chang, H.-I.; Davies, N.M.; McMillan, N.A.J. Development of a Novel Method for Formulating Stable SiRNA-Loaded Lipid Particles for in Vivo Use. Pharm. Res. 2009, 26, 512–522.
- McCaskill, J.; Singhania, R.; Burgess, M.; Allavena, R.; Wu, S.; Blumenthal, A.; McMillan, N.A. Efficient Biodistribution and Gene Silencing in the Lung Epithelium via Intravenous Liposomal Delivery of SiRNA. Mol. Ther. Nucleic Acids 2013, 2, e96.
- Khaitov, M.; Nikonova, A.; Shilovskiy, I.; Kozhikhova, K.; Kofiadi, I.; Vishnyakova, L.; Nikolskii, A.; Gattinger, P.; Kovchina, V.; Barvinskaia, E.; et al. Silencing of SARS-CoV-2 with Modified SiRNA-Peptide Dendrimer Formulation. Allergy 2021, 76, 2840–2854.
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of MiR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420.
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. SiRNA Versus MiRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252.
- Doench, J.G.; Sharp, P.A. Specificity of MicroRNA Target Selection in Translational Repression. Genes Dev. 2004, 18, 504–511.
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. SiRNA: Mechanism of Action, Challenges, and Therapeutic Approaches. Eur. J. Pharmacol. 2021, 905, 174178.
- Hennig, T.; Prusty, A.B.; Kaufer, B.B.; Whisnant, A.W.; Lodha, M.; Enders, A.; Thomas, J.; Kasimir, F.; Grothey, A.; Klein, T.; et al. Selective Inhibition of MiRNA Processing by a Herpesvirus-Encoded MiRNA. Nature 2022, 605, 539–544.
- Gahlawat, A.W.; Witte, T.; Haarhuis, L.; Schott, S. A Novel Circulating MiRNA Panel for Non-Invasive Ovarian Cancer Diagnosis and Prognosis. Br. J. Cancer 2022. online ahead of print.
- Strand, S.H.; Schmidt, L.; Weiss, S.; Borre, M.; Kristensen, H.; Rasmussen, A.K.I.; Daugaard, T.F.; Kristensen, G.; Stroomberg, H.V.; Røder, M.A.; et al. Validation of the Four-MiRNA Biomarker Panel MiCaP for Prediction of Long-Term Prostate Cancer Outcome. Sci. Rep. 2020, 10, 10704.
- Li, Q.; Lowey, B.; Sodroski, C.; Krishnamurthy, S.; Alao, H.; Cha, H.; Chiu, S.; El-Diwany, R.; Ghany, M.G.; Liang, T.J. Cellular MicroRNA Networks Regulate Host Dependency of Hepatitis C Virus Infection. Nat. Commun. 2017, 8, 1789.
- Moffett, H.F.; Cartwright, A.N.R.; Kim, H.-J.; Godec, J.; Pyrdol, J.; Äijö, T.; Martinez, G.J.; Rao, A.; Lu, J.; Golub, T.R.; et al. The MicroRNA MiR-31 Inhibits CD8+ T Cell Function in Chronic Viral Infection. Nat. Immunol. 2017, 18, 791–799.
- Li, C.; Hu, X.; Li, L.; Li, J.-H. Differential MicroRNA Expression in the Peripheral Blood from Human Patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590.
- Samy, A.; Maher, M.A.; Abdelsalam, N.A.; Badr, E. SARS-CoV-2 Potential Drugs, Drug Targets, and Biomarkers: A Viral-Host Interaction Network-Based Analysis. Sci. Rep. 2022, 12, 11934.
- Khan, A.-A.-K.; Sany, R.U.; Islam, S.; Islam, A.B.M.K. Epigenetic Regulator MiRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Front. Genet. 2020, 11, 765.
- Panda, M.; Kalita, E.; Singh, S.; Kumar, K.; Rao, A.; Prajapati, V.K. MiRNA-SARS-CoV-2 Dialogue and Prospective Anti-COVID-19 Therapies. Life Sci. 2022, 305, 120761.
- Ahmed, J.Q.; Maulud, S.Q.; Dhawan, M.; Priyanka; Choudhary, O.P.; Jalal, P.J.; Ali, R.K.; Tayib, G.A.; Hasan, D.A. MicroRNAs in the Development of Potential Therapeutic Targets against COVID-19: A Narrative Review. J. Infect. Public Health 2022, 15, 788–799.
- Vitravene Study Group. A Randomized Controlled Clinical Trial of Intravitreous Fomivirsen for Treatment of Newly Diagnosed Peripheral Cytomegalovirus Retinitis in Patients with Aids. Am. J. Ophthalmol. 2002, 133, 467–474.
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694.
- Ren, H.; Zhang, Z.; Zhang, W.; Feng, X.; Xu, L. Prodrug-Type Antisense Oligonucleotides with Enhanced Nuclease Stability and Anti-Tumour Effects. Eur. J. Pharm. Sci. 2021, 162, 105832.
- Kole, R. RNA Therapeutics: Beyond RNA Interference and Antisense Oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140.
- DeVos, S.L.; Miller, T.M. Antisense Oligonucleotides: Treating Neurodegeneration at the Level of RNA. Neurotherapeutics 2013, 10, 486–497.
- Le, B.T.; Raguraman, P.; Kosbar, T.R.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Antisense Oligonucleotides Targeting Angiogenic Factors as Potential Cancer Therapeutics. Mol. Ther. Nucleic Acids 2019, 14, 142–157.
- Berber, B.; Aydin, C.; Kocabas, F.; Guney-Esken, G.; Yilancioglu, K.; Karadag-Alpaslan, M.; Caliseki, M.; Yuce, M.; Demir, S.; Tastan, C. Gene Editing and RNAi Approaches for COVID-19 Diagnostics and Therapeutics. Gene Ther. 2020, 28, 290–305.
- Crooke, S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77.
- Lim, K.H.; Han, Z.; Jeon, H.Y.; Kach, J.; Jing, E.; Weyn-Vanhentenryck, S.; Downs, M.; Corrionero, A.; Oh, R.; Scharner, J.; et al. Antisense Oligonucleotide Modulation of Non-Productive Alternative Splicing Upregulates Gene Expression. Nat. Commun. 2020, 11, 3501.
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-Length Transcript Characterization of SF3B1 Mutation in Chronic Lymphocytic Leukemia Reveals Downregulation of Retained Introns. Nat. Commun. 2020, 11, 1438.
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027.
- Gouni-Berthold, I. The Role of Antisense Oligonucleotide Therapy against Apolipoprotein-CIII in Hypertriglyceridemia. Atheroscler. Suppl. 2017, 30, 19–27.
- Zorzi, F.; Angelucci, E.; Sedda, S.; Pallone, F.; Monteleone, G. Smad7 Antisense Oligonucleotide-Based Therapy for Inflammatory Bowel Diseases. Dig. Liver Dis. 2013, 45, 552–555.
- Dulla, K.; Slijkerman, R.; van Diepen, H.C.; Albert, S.; Dona, M.; Beumer, W.; Turunen, J.J.; Chan, H.L.; Schulkens, I.A.; Vorthoren, L.; et al. Antisense Oligonucleotide-Based Treatment of Retinitis Pigmentosa Caused by USH2A Exon 13 Mutations. Mol. Ther. 2021, 29, 2441–2455.
- Yuen, M.-F.; Gane, E.; Kim, D.J.; Chan, H.; Surujbally, B.; Pavlovic, V.; Triyatni, M.; Grippo, J.; Kim, H.J.; Leerapun, A.; et al. RO7062931 Antisense Oligonucleotide Phase 1 Study Demonstrates Target Engagement in Patients with Chronic Hepatitis B on Established Nucleos(t)Ide Therapy. J. Hepatol. 2020, 73, S51.
- Billioud, G.; Kruse, R.L.; Carrillo, M.; Whitten-Bauer, C.; Gao, D.; Kim, A.; Chen, L.; McCaleb, M.L.; Crosby, J.R.; Hamatake, R.; et al. In Vivo Reduction of Hepatitis B Virus Antigenemia and Viremia by Antisense Oligonucleotides. J. Hepatol. 2016, 64, 781–789.
- Chery, J.; Petri, A.; Wagschal, A.; Lim, S.-Y.; Cunningham, J.; Vasudevan, S.; Kauppinen, S.; Näär, A.M. Development of Locked Nucleic Acid Antisense Oligonucleotides Targeting Ebola Viral Proteins and Host Factor Niemann-Pick C1. Nucleic Acid Ther. 2018, 28, 273–284.
- Lenartowicz, E.; Nogales, A.; Kierzek, E.; Kierzek, R.; Martínez-Sobrido, L.; Turner, D.H. Antisense Oligonucleotides Targeting Influenza A Segment 8 Genomic RNA Inhibit Viral Replication. Nucleic Acid Ther. 2016, 26, 277–285.
- Deas, T.S.; Bennett, C.J.; Jones, S.A.; Tilgner, M.; Ren, P.; Behr, M.J.; Stein, D.A.; Iversen, P.L.; Kramer, L.D.; Bernard, K.A.; et al. In Vitro Resistance Selection and In Vivo Efficacy of Morpholino Oligomers against West Nile Virus. Antimicrob. Agents Chemother. 2007, 51, 2470–2482.
- Sun, L.; Li, P.; Ju, X.; Rao, J.; Huang, W.; Ren, L.; Zhang, S.; Xiong, T.; Xu, K.; Zhou, X.; et al. In Vivo Structural Characterization of the SARS-CoV-2 RNA Genome Identifies Host Proteins Vulnerable to Repurposed Drugs. Cell 2021, 184, 1865–1883.e20.
- Hasan, M.M.; Ragnarsson, L.; Cardoso, F.C.; Lewis, R.J. Transfection Methods for High-Throughput Cellular Assays of Voltage-Gated Calcium and Sodium Channels Involved in Pain. PLoS ONE 2021, 16, e0243645.
- Clark, D.P.; Pazdernik, N.J.; McGehee, M.R. Manipulation of Nucleic Acids. In Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 132–166. ISBN 978-0-12-813288-3.
- Ahn, D.-G.; Lee, W.; Choi, J.-K.; Kim, S.-J.; Plant, E.P.; Almazán, F.; Taylor, D.R.; Enjuanes, L.; Oh, J.-W. Interference of Ribosomal Frameshifting by Antisense Peptide Nucleic Acids Suppresses SARS Coronavirus Replication. Antivir. Res. 2011, 91, 1–10.
- Kelly, J.A.; Woodside, M.T.; Dinman, J.D. Programmed −1 Ribosomal Frameshifting in Coronaviruses: A Therapeutic Target. Virology 2021, 554, 75–82.
- Jacobsen, L.B.; Calvin, S.A.; Colvin, K.E.; Wright, M. FuGENE 6 Transfection Reagent: The Gentle Power. Methods 2004, 33, 104–112.
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259.
- Workman, R.E.; Pammi, T.; Nguyen, B.T.K.; Graeff, L.W.; Smith, E.; Sebald, S.M.; Stoltzfus, M.J.; Euler, C.W.; Modell, J.W. A Natural Single-Guide RNA Repurposes Cas9 to Autoregulate CRISPR-Cas Expression. Cell 2021, 184, 675–688.e19.
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712.
- Barrangou, R.; van der Oost, J. (Eds.) CRISPR-Cas Systems; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-642-34656-9.
- Sioud, M. (Ed.) RNA Interference and CRISPR Technologies: Technical Advances and New Therapeutic Opportunities (Methods in Molecular Biology); Springer: New York, NY, USA, 2020; Volume 2115, ISBN 978-1-07-160289-8.
- Soares, F.; Chen, B.; Lee, J.B.; Ahmed, M.; Ly, D.; Tin, E.; Kang, H.; Zeng, Y.; Akhtar, N.; Minden, M.D.; et al. CRISPR Screen Identifies Genes That Sensitize AML Cells to Double-Negative T-Cell Therapy. Blood 2021, 137, 2171–2181.
- Zeballos, C.M.A.; Gaj, T. Next-Generation CRISPR Technologies and Their Applications in Gene and Cell Therapy. Trends Biotechnol. 2021, 39, 692–705.
- Liao, H.-K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.-J.; et al. Use of the CRISPR/Cas9 System as an Intracellular Defense against HIV-1 Infection in Human Cells. Nat. Commun. 2015, 6, 6413.
- Yin, D.; Ling, S.; Wang, D.; Dai, Y.; Jiang, H.; Zhou, X.; Paludan, S.R.; Hong, J.; Cai, Y. Targeting Herpes Simplex Virus with CRISPR–Cas9 Cures Herpetic Stromal Keratitis in Mice. Nat. Biotechnol. 2021, 39, 567–577.
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 2017, 37, 67–78.
- Méndez-Mancilla, A.; Wessels, H.-H.; Legut, M.; Kadina, A.; Mabuchi, M.; Walker, J.; Robb, G.B.; Holden, K.; Sanjana, N.E. Chemically Modified Guide RNAs Enhance CRISPR-Cas13 Knockdown in Human Cells. Cell Chem. Biol. 2021, 29, 321–327.
- Smargon, A.A.; Shi, Y.J.; Yeo, G.W. RNA-Targeting CRISPR Systems from Metagenomic Discovery to Transcriptomic Engineering. Nat. Cell Biol. 2020, 22, 143–150.
- Yin, L.; Zhao, F.; Sun, H.; Wang, Z.; Huang, Y.; Zhu, W.; Xu, F.; Mei, S.; Liu, X.; Zhang, D.; et al. CRISPR-Cas13a Inhibits HIV-1 Infection. Mol. Ther. Nucleic Acids 2020, 21, 147–155.
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876.e12.
- Fareh, M.; Zhao, W.; Hu, W.; Casan, J.M.L.; Kumar, A.; Symons, J.; Zerbato, J.M.; Fong, D.; Voskoboinik, I.; Ekert, P.G.; et al. Reprogrammed CRISPR-Cas13b Suppresses SARS-CoV-2 Replication and Circumvents Its Mutational Escape through Mismatch Tolerance. Nat. Commun. 2021, 12, 4270.
- Dufait, I.; Liechtenstein, T.; Lanna, A.; Bricogne, C.; Laranga, R.; Padella, A.; Breckpot, K.; Escors, D. Retroviral and Lentiviral Vectors for the Induction of Immunological Tolerance. Scientifica 2012, 2012, 694137.
- Behr, M.; Zhou, J.; Xu, B.; Zhang, H. In Vivo Delivery of CRISPR-Cas9 Therapeutics: Progress and Challenges. Acta Pharm. Sin. B 2021, 11, 2150–2171.
- Blanchard, E.L.; Vanover, D.; Bawage, S.S.; Tiwari, P.M.; Rotolo, L.; Beyersdorf, J.; Peck, H.E.; Bruno, N.C.; Hincapie, R.; Michel, F.; et al. Treatment of Influenza and SARS-CoV-2 Infections via MRNA-Encoded Cas13a in Rodents. Nat. Biotechnol. 2021, 39, 717–726.
- Patel, A.K.; Kaczmarek, J.C.; Bose, S.; Kauffman, K.J.; Mir, F.; Heartlein, M.W.; DeRosa, F.; Langer, R.; Anderson, D.G. Inhaled Nanoformulated MRNA Polyplexes for Protein Production in Lung Epithelium. Adv. Mater. 2019, 31, 1805116.
More
