Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 4 by Jessie Wu and Version 4 by Jessie Wu.
Photolyase is a protein that has various functions, among which is the repair of DNA damaged from exposure to UV rays from the sun.
- Photolyase
- DNA Repair
- Enzyme
- Topical applications
1. Introduction
The presence of photolyases has been observed in fish, amphibians, birds, and a few marsupials [1]; nevertheless, in higher plants and animals, the ability to repair DNA was lost during evolution (Figure 1). Therefore, their function is limited to regulating growth and acting as blue-light photoreceptors; these enzymes are known as cryptochromes [2].
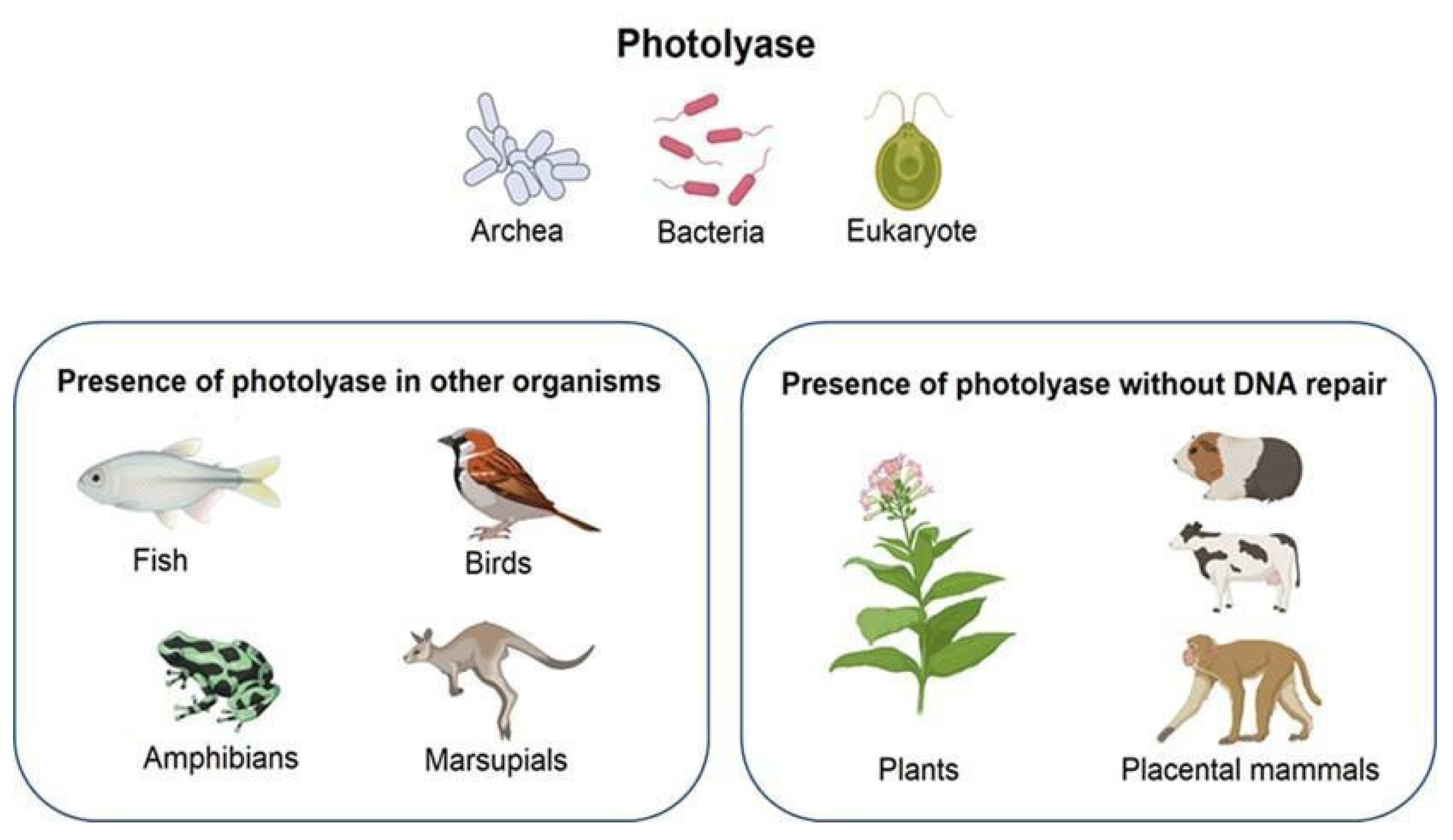
Figure 1. Presence of photolyase in different organisms. Due to evolution, some species lost the ability to repair DNA. Created with BioRender.com.
The first precedent of enzymes with photolyase-like activity was discovered in 1993 in a plant of the Brassicaceae family native to Europe, Arabidopsis thaliana [3]. In other plants such as white mustard (Sinapis alba), the same photolyase cofactors are present; however, these plants lack DNA repair activity [4]. However, the photoreactivation process and DNA repair with photolyase are available and have been demonstrated in Streptomyces griseus [5] and bacteriophages [6].
In cyanobacteria, the existence of photolyase is reported in Anacystis nidulans [7]. The activity of photolyase enzymes has been reported mainly in environments with high exposure to UV rays, such as the case of twelve species of diatoms from Antarctica that demonstrated a DNA repair response to UV radiation damage [8].
In eukaryotic organisms, the Antarctic alga Chlamydomonas sp. ICE-L, which is developed in high-irradiation environments, can use mechanisms that reduce UV radiation damage [9].
2. Type of Enzyme
Photolyases are monomeric proteins with a molecular mass from 50 to 61 kDa. They are made up of 450–550 amino acids and two unsorted covalently bound chromophores as cofactors. One of the cofactors is always flavin adenine dinucleotide FAD, and the second is methenyltetrahydrofolate (MTHF) or 8-hydroxy-7, 8-didemethyl-5-deazariboflavin (8-HDF) [10]. Light is an indispensable resource in the photoreactivation process for the conversion of enzyme–substrate complexes into additional DNA repair products [11]. The surface of photolyases is characterized by a positive charge near the substrate binding which promotes the interaction with DNA [12]. The structure of photolyase consists of two domains: a C-terminal α-helical catalytic domain that contains the flavin cofactor and an N-terminal α/β domain [13].
The superfamily of chromophores/photolyases (CRY/PHR) consists of subfamilies (Figure 2) [14][15], of which there are three types of photolyases that have been identified to repair a specific type of dimer: (1) CPD photolyase, responsible for repairing CPD, (6-4); (2) photolyase, which repairs (6-4) pyrimidine pyrimidone; and (3) cryptochrome-DASH, which causes a variety of physiological changes to DNA [16][17].
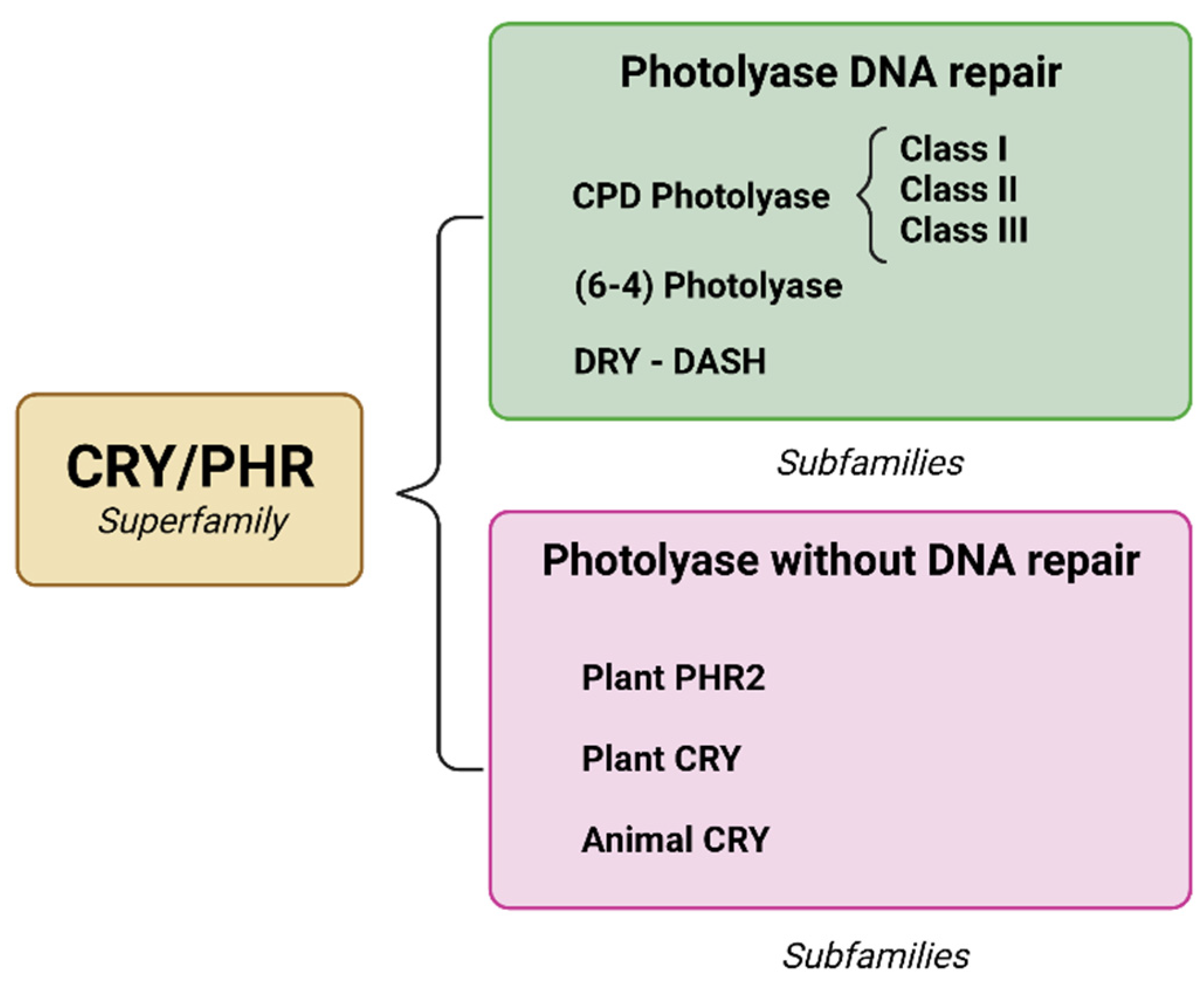
Figure 2. Integration of the CRY/PHR superfamily and the emergence of subfamilies due to evolutionary changes. The main distinctions are the ability to repair specific DNA and loss of photorepair capacity. Created with BioRender.com.
3. Photolyase and Microorganisms
Photolyase biosynthesis begins with the transfer of an electron from the anionic chromophore FADH−, promoted by light. In a catalytically active form, this binds to damaged DNA in a high-affinity, light-independent step [18][19].
To study numerous characteristics of the photolyases obtained from various organisms such as structural, physical, and mechanical properties, researchers have employed genetic manipulation approaches to promote the overexpression of DNA repair enzymes, since biosynthesis naturally provides low concentrations. Subsequently, extraction and purification processes are performed to obtain better quality enzymes (Table 1) [20]. After these steps, generally, between 15 and 25 mg of photolyase with a purity greater than 98% is obtained. The quality of the enzyme obtained is reflected in the color of the extract: a dark blue color indicates a good quality product. Therefore, it is important to measure and quantify the absorbance spectrum after enzyme purification [20].
Table 1. Studies demonstrating the presence of different types of photolyases in various organisms and the extraction and purification methods necessary to prove their DNA repair activity.
| Microorganisms | Genus | Type Photolyase | Extraction and Purification | References |
|---|---|---|---|---|
| Agrobacterium fabrum | Prokaryote | (6-4) Photolyase | Heated and cleared by centrifugation HPLC column from Macherey and Nagel | [21] |
| Rhodococcus sp. NJ-530 | Marine bacterium | CPD Class I | Disrupted with ultrasonication, Ni-NTA resin | [22] |
| Chlamydomonas sp. ICE-L | Psychrophilic microalga | (6-4) Photolyase | Disrupted with ultrasonication, Ni-NTA resin | [23] |
| Hymenobacter sp. | Antarctic bacterium | CPD Class I | Lysed with sonication, Ni-NTA resin | [24] |
| Methanosarcina mazei Mm0852 | Archaea | CPD Class II | Cell disruption with lysozyme, EDTA and PMSF with an emulsifier, Ni-NTA resin | [25] |
| Pohlia nutans M211 | Antarctic Moss | CPD Class II and (6-4) Photolyase | Ultrasonic cell disruptor, Ni-NTA resin | [26] |
| Phaeodactylum tricornutum ICE-H | Antarctic diatom | CPD Class II | Ultrasonic cell disruptor, Ni-NTA resin | [27] |
| Caulobacter crescentus | Oligotrophic bacterium | CPD Class III | Heated and cleared with centrifugation, purified by affinity chromatography on amylose resin | [28] |
| Mucor circinelloides | Fungus | CRY-DASH | Disrupted with a French press, affinity chromatography-HisTrap HP column | [29] |
| Phycomyces blakesleeanus (NRRL1555) | Fungus | CRY-DASH | Disrupted with a French press, affinity chromatography-His Trap HP column | [30] |
4. DNA Damage by UV Irradiation
The organisms and cells that inhabit the earth naturally are continuously exposed to genotoxic agents present in the environment. Sunlight, as a source of UV rays, is one of the main genotoxic agents; nonetheless, it is essential for the development of life, for example, in the process of photosynthesis [31].
The damage caused by exposure to UV radiation can trigger various skin reactions, mainly (as mentioned before) by affecting pyrimidine dimers; erythema, immunosuppression, and melanogenesis are just some of the disorders that can occur. Hence, it has been proven that overexposure to sunlight almost irreversibly damages skin cells [32].
5. Photolyase Mechanism of Action
There are multiple mechanisms of DNA repair: direct reversal [33], base excision, nucleotide excision [34], mismatch [35], single-strand break, and double-strand break repair [36].
Photolyases are light-driven DNA repair enzymes which function specifically in the reversal of genomic lesions induced by UV radiation [37]. An important DNA repair mechanism for mutagenic and cytotoxic UV-induced photolesions in DNA is photoreactivation, which utilizes enzyme photolyase for reverting modified nitrogenous bases into normal form, employing blue wavelength [38]. DNA repair to minimize mutagenic changes is divided into two main mechanisms: single-strand (ss) and double-strand (ds) DNA damage repair. In the same way, ss and ds DNA repair are divided into direct reversal repair, nucleotide excision repair, base excision repair, and mismatch repair for ss DNA repair and homologous recombination and non-homologous end-joining repairs for ds DNA repairs [39].
The photolyase for the CPD and 6-4PP lesions can be divided into CPD photolyases based on the photoproduct that they recognize, which are subdivided in relation to the amino acid sequence they possess, and 6-4PP photolyases, respectively [40]. Photolyases can possess flavin adenine dinucleotide (FAD) in four different redox states: oxidized (FAD), anionic semiquinone (FAD−), neutral semiquinone (FADH), and anionic hydroquinone (FADH−) [39]. Different redox states will act differently in the absorption spectrum. While FAD and FAD− mainly absorb UV-A, and blue light, FADH absorbs blue, green, and red light, and FADH− does not absorb visible light. An absorption spectrum was determined from two organisms by following flavin intermediates during the catalytic process, showing the mode of action on the DNA repair by photolyase based on the absorption properties of FAD [41].
According to Wang et al. [41], DNA repair by photolyase can be divided into three steps: (i) Recognition, which is a light-independent process where CPD or (6-4) photoproduct in the damaged DNA forms a photolyase/DNA complex by flipping into the active site containing the flavin cofactor of the DNA photolyase. (ii) During the catalytic reaction [42] taking place when FAD is at a fully reduced state (FADH−), the methenytetrahydrofolate (MTHF), a photolyase chromophore, transfers energy to FADH− through the absorption of photons from the blue-light spectrum, changing FADH− to an exciting form of FADH−. In this step, CPD or the (6-4) photoproduct ring is opened by bond dissociation in the dimer radical anion, and electron transfers from FADH− to the lesions occur [39][43]. (iii) Finally, the separation step completes the repair process by the departure of repaired DNA from photolyases.
As previously mentioned, FAD oxidation states influence the light absorption spectra; hence, catalytic reactions for DNA repair from FADH− take place under blue-light conditions. Considering the poor permeability of living tissues to blue light, works that aim to remove the barrier of this range of light have been developed using harvesting or antenna chromophores. Antenna chromophores are utilized by some photolyase to gain the photoreception ability of a light range, and the development and application of artificial antenna chromophores demonstrate an increase of up to 1.5-fold in DNA repair activity. This is a promising strategy area for the optimization of photolyase DNA repair [43]. Nevertheless, reports on the application of artificial antenna chromophores remain scarce.
6. Immobilization and Biocarriers
Photolyase offers outstanding protection and recovery mechanisms against sunlight-induced DNA damage, but if the enzyme lacks the capacity of reaching the site where it should intervene, or lacks the stability necessary to complete this, its utilization will go to waste.
Currently, immobilization strategies such as the encapsulation of photolyases within liposomes are the most used method to stabilize and deliver the protein; nonetheless, other methods, such as the application of nanomaterials, are being explored [40]. There are an assortment of technologies and systems currently being used as an approach to deliver active ingredients in skincare, such as antioxidants that can be useful and might offer an alternative to liposomes. Some of these technologies include the use of different formulations, such as gels, hydrogels, and emulsions (micro and nano-emulsions), the usage of other vesicular delivery systems instead of liposomes, such as ethosomes, transfersomes, niosomes, and non-vesicular particles such as solid lipid nanoparticles, and nanostructured lipid carriers, polymeric nanoparticles, nanocrystals, and—lastly—carbon, metal, or metal oxide nanoparticles. These immobilization technologies seek to improve long-term stability and sensitivity in order to provide a better catalytic activity; however, it is necessary to explore their efficiency according to their final applications [44][45][46][47][48][49][50][51].
References
- Menck, C.F.M. Shining a light on photolyases. Nat. Genet. 2002, 32, 338–339.
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.O.; Van Der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011, 62, 335–364.
- Ahmad, M.; Cashmore, A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166.
- Malhotra, K.; Kim, S.T.; Batschauer, A.; Dawut, L.; Sancar, A. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 1995, 34, 6892–6899.
- Kelner, A. Effect of Visible Light on the Recovery of Streptomyces Griseus Conidia from Ultra-violet Irradiation Injury. Proc. Natl. Acad. Sci. USA 1949, 35, 73–79.
- Dulbecco, R. Reactivation of ultra-violet-inactivated bacteriophage by visible light. Nature 1949, 163, 949–950.
- Aubert, C.; Mathis, P.; Eker, A.P.M.; Brettel, K. Intraprotein electron transfer between tyrosine and tryptophan in DNA photolyase from Anacystis nidulans. Proc. Natl. Acad. Sci. USA 1999, 96, 5423–5427.
- Karentz, D.; Cleaver, J.E.; Mitchell, D.L. Cell survival characteristics and molecular responses of Antarctic phytoplankton to ultraviolet-B radiation. J. Phycol. 1991, 27, 326–341.
- Li, C.; Ma, L.; Mou, S.; Wang, Y.; Zheng, Z.; Liu, F.; Qi, X.; An, M.; Chen, H.; Miao, J. Cyclobutane pyrimidine dimers photolyase from extremophilic microalga: Remarkable UVB resistance and efficient DNA damage repair. Mutat. Res. 2015, 773, 37–42.
- Sancar, A. Structure and Function of DNA Photolyase and Cryptochrome Blue-Light Photoreceptors. Chem. Rev. 2003, 103, 2203–2238.
- Sancar, G.B. DNA photolyases: Physical properties, action mechanism, and roles in dark repair. Mutat. Res. 1990, 236, 147–160.
- Müller, M.; Carell, T. Structural biology of DNA photolyases and cryptochromes. Curr. Opin. Struct. Biol. 2009, 19, 277–285.
- Zhang, M.; Wang, L.; Zhong, D. Photolyase: Dynamics and Mechanisms of Repair of Sun-Induced DNA Damage. Photochem. Photobiol. 2017, 93, 78–92.
- Mei, Q.; Dvornyk, V. Evolutionary History of the Photolyase/Cryptochrome Superfamily in Eukaryotes. PLoS ONE 2015, 10, e0135940.
- Öztürk, N.; Song, S.H.; Özgür, S.; Selby, C.P.; Morrison, L.; Partch, C.; Zhong, D.; Sancar, A. Structure and function of animal cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 119–131.
- Selby, C.P.; Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17696–17700.
- Essen, L.O.; Klar, T. Light-driven DNA repair by photolyases. Cell. Mol. Life Sci. 2006, 63, 1266–1277.
- Graf, D.; Wesslowski, J.; Ma, H.; Scheerer, P.; Krauß, N.; Oberpichler, I.; Zhang, F.; Lamparter, T. Key Amino Acids in the Bacterial (6-4) Photolyase PhrB from Agrobacterium fabrum. PLoS ONE 2015, 10, e0140955.
- He, Y.; Qu, C.; Zhang, L.; Miao, J. DNA photolyase from Antarctic marine bacterium Rhodococcus sp. NJ-530 can repair DNA damage caused by ultraviolet. 3 Biotech 2021, 11, 102.
- An, M.; Zheng, Z.; Qu, C.; Wang, X.; Chen, H.; Shi, C.; Miao, J. The first (6-4) photolyase with DNA damage repair activity from the Antarctic microalga Chlamydomonas sp. ICE-L. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2018, 809, 13–19.
- Marizcurrena, J.J.; Martínez-López, W.; Ma, H.; Lamparter, T.; Castro-Sowinski, S. A highly efficient and cost-effective recombinant production of a bacterial photolyase from the Antarctic isolate Hymenobacter sp. UV11. Extremophiles 2019, 23, 49–57.
- Kiontke, S.; Geisselbrecht, Y.; Pokorny, R.; Carell, T.; Batschauer, A.; Essen, L.O. Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J. 2011, 30, 4437–4449.
- An, M.; Qu, C.; Miao, J.; Sha, Z. A Class II CPD Photolyase and a 6-4 Photolyase with Photorepair Activity from the Antarctic Moss Pohlia nutans M211. Photochem. Photobiol. 2021, 97, 1527–1533.
- An, M.; Qu, C.; Miao, J.; Sha, Z. Two class II CPD photolyases, PiPhr1 and PiPhr2, with CPD repair activity from the Antarctic diatom Phaeodactylum tricornutum ICE-H. 3 Biotech 2021, 11, 377.
- Öztürk, N.; Kao, Y.T.; Selby, C.P.; Kavakli, I.H.; Partch, C.L.; Zhong, D.; Sancar, A. Purification and Characterization of a Type III Photolyase from Caulobacter crescentus. Biochemistry 2008, 47, 10255.
- Navarro, E.; Niemann, N.; Kock, D.; Dadaeva, T.; Gutiérrez, G.; Engelsdorf, T.; Kiontke, S.; Corrochano, L.M.; Batschauer, A.; Garre, V. The DASH-type Cryptochrome from the Fungus Mucor circinelloides Is a Canonical CPD-Photolyase. Curr. Biol. 2020, 30, 4483–4490.e4.
- Tagua, V.G.; Pausch, M.; Eckel, M.; Gutiérrez, G.; Miralles-Durán, A.; Sanz, C.; Eslava, A.P.; Pokorny, R.; Corrochano, L.M.; Batschauer, A. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 15130–15135.
- Schleicher, E.; Heßling, B.; Illarionova, V.; Bacher, A.; Weber, S.; Richter, G.; Gerwert, K. Light-induced reactions of Escherichia coli DNA photolyase monitored by Fourier transform infrared spectroscopy. FEBS J. 2005, 272, 1855–1866.
- Brash, D.E.; Franklin, W.A.; Sancar, G.B.; Haseltine, W.A. Escherichia coli DNA photolyase reverses cyclobutane pyrimidine dimers but not pyrimidine-pyrimidone (6-4) photoproducts. J. Biol. Chem. 1985, 260, 11438–11441.
- Sancar, G.B.; Sancar, A. Purification and characterization of DNA photolyases. Methods Enzymol. 2006, 408, 121–156.
- Mullenders, L.H.F. Solar UV damage to cellular DNA: From mechanisms to biological effects. Photochem. Photobiol. Sci. 2018, 17, 1842–1852.
- Marrot, L.; Meunier, J.R. Skin DNA photodamage and its biological consequences. J. Am. Acad. Dermatol. 2008, 58, S139–S148.
- Ahmad, A.; Nay, S.L.; O’Connor, T.R. Direct Reversal Repair in Mammalian Cells. In Advances in DNA Repair; Chen, C., Ed.; InTech: Rijeka, Croatia, 2015.
- Petruseva, I.O.; Evdokimov, A.N.; Lavrik, O.I. Molecular mechanism of global genome nucleotide excision repair. Acta Nat. 2014, 6, 23–34.
- Fukui, K. DNA Mismatch Repair in Eukaryotes and Bacteria. J. Nucleic Acids 2010, 2010, 260512.
- Vítor, A.C.; Huertas, P.; Legube, G.; de Almeida, S.F. Studying DNA Double-Strand Break Repair: An Ever-Growing Toolbox. Front. Mol. Biosci. 2020, 7, 24.
- Huang, R.; Zhou, P.K. DNA Damage Repair: Historical Perspectives, Mechanistic Pathways and Clinical Translation for Targeted Cancer Therapy; Springer: Greer, SA, USA, 2021; Volume 6, ISBN 4139202100648.
- Pathak, J.; Rajneesh; Singh, P.R.; Häder, D.P.; Sinha, R.P. UV-induced DNA damage and repair: A cyanobacterial perspective. Plant Gene 2019, 19, 100194.
- Kavakli, I.H.; Ozturk, N.; Gul, S. DNA repair by photolyases. Adv. Protein Chem. Struct. Biol. 2019, 115, 1–19.
- Ramírez, N.; Serey, M.; Illanes, A.; Piumetti, M.; Ottone, C. Immobilization strategies of photolyases: Challenges and perspectives for DNA repairing application. J. Photochem. Photobiol. B Biol. 2021, 215, 112113.
- Wang, J.; Du, X.; Pan, W.; Wang, X.; Wu, W. Photoactivation of the cryptochrome/photolyase superfamily. J. Photochem. Photobiol. C Photochem. Rev. 2015, 22, 84–102.
- Brettel, K.; Müller, P.; Yamamoto, J. Kinetics of Electron Returns in Successive Two-Photon DNA Repair by (6-4) Photolyase. ACS Catal. 2022, 12, 3041–3045.
- Terai, Y.; Sato, R.; Matsumura, R.; Iwai, S.; Yamamoto, J. Enhanced DNA repair by DNA photolyase bearing an artificial light-harvesting chromophore. Nucleic Acids Res. 2020, 48, 10076–10086.
- Benson, H.A.E.; Grice, J.E.; Mohammed, Y.; Namjoshi, S.; Roberts, M.S. Topical and Transdermal Drug Delivery: From Simple Potions to Smart Technologies. Curr. Drug Deliv. 2019, 16, 444.
- Pavlou, P.; Siamidi, A.; Varvaresou, A.; Vlachou, M. Skin Care Formulations and Lipid Carriers as Skin Moisturizing Agents. Cosmetics 2021, 8, 89.
- Roberts, M.S.; Cheruvu, H.S.; Mangion, S.E.; Alinaghi, A.; Benson, H.A.E.; Mohammed, Y.; Holmes, A.; van der Hoek, J.; Pastore, M.; Grice, J.E. Topical drug delivery: History, percutaneous absorption, and product development. Adv. Drug Deliv. Rev. 2021, 177, 113929.
- Eskens, O.; Amin, S. Challenges and effective routes for formulating and delivery of epidermal growth factors in skin care. Int. J. Cosmet. Sci. 2021, 43, 123–130.
- Salvioni, L.; Morelli, L.; Ochoa, E.; Labra, M.; Fiandra, L.; Palugan, L.; Prosperi, D.; Colombo, M. The emerging role of nanotechnology in skincare. Adv. Colloid Interface Sci. 2021, 293, 102437.
- Montenegro, L. Nanocarriers for skin delivery of cosmetic antioxidants. J. Pharm. Pharmacogn. Res. 2014, 2, 73–92.
- Kim, B.; Cho, H.-E.; Moon, S.H.; Ahn, H.-J.; Bae, S.; Cho, H.-D.; An, S. Transdermal delivery systems in cosmetics. Biomed. Dermatol. 2020, 4, 10.
- Travis, B.; Darren, S.; Zimei, W. Pharmaceutical Strategies for the Topical Dermal Delivery of Peptides/Proteins for Cosmetic and Therapeutic Applications. Austin J. Pharmacol. Ther. 2014, 2, 1–10.
More
