PAX5, a member of the Paired Box (PAX) transcription factor family, is an essential factor for B-lineage identity during lymphoid differentiation. Mechanistically, PAX5 controls gene expression profiles, pivotal to cellular processes such as viability, proliferation, and differentiation. Given its crucial function in B-cell development, PAX5 aberrant expression also correlates with hallmark cancer processes leading to hematological and other types of cancer lesion.
- PAX5
- BSAP
- B-cell
- differentiation
- transcription factor
1. Introduction
The Paired Box (PAX) gene family encodes nine transcription factors (PAX1–9), which regulate gene expression programs in tissue development[1]. Although PAX transcription factors share a highly similar paired-box DNA-binding domain, they are classified into four subgroups (I–IV) based on additional functional domains such as the octapeptide and the homeodomain, which are generally located in the protein’s internal and amino-terminal regions respectively[2][3]. Given their structural resemblance, PAX members from a particular subgroup account for similar activities and functions. For example, PAX genes in subgroups II (PAX2, PAX5, and PAX8) and III (PAX3 and PAX7) are commonly involved in processes including cell survival, motility, and tumor progression. Conversely, members from subgroup I (PAX1 and PAX9) and IV (PAX4 and PAX6) seem less involved in cancer processes[4]. The expression of PAX family gene products is also generally tissue specific. For instance, PAX2 expression has been described in kidney and optic nerve development[5], whereas PAX5 has mostly been associated with the development of the central nervous system, of B-lymphocytes, and spermatogenesis[6]. Furthermore, the expression patterns of subgroup II members are reported to be altered in various cancer tissues, which suggests a distinctive role for these PAX gene products in the regulation of specific malignancies[1]. Amongst these members, PAX5 has been extensively studied and characterized for its role in cancer pathogenesis.
2. Expression and Tissue Specificity
The human PAX5 gene locus is located on the 9p13 chromosomal region known to undergo a high degree of alterations leading to its implication in cancer development and progression[7]. Structurally, the PAX5 gene is characterized by two known distinct promoters, resulting in two alternative transcriptional initiation sites known as PAX5A and PAX5B[8]. Both transcripts share the same sequence encoded by exon 2 through exon 10. However, they have different sequences in their first exon (1A or 1B), which is dependently linked to their respective promoter regions (PAX5 1A versus PAX5 1B). Both PAX5A and PAX5B protein variants consist of a 52-kD protein known as the B-cell lineage-Specific Activator Protein (BSAP), which was initially identified as an essential regulator of early B-cell differentiation and commitment[6][9]. Despite their structural similarities, PAX5A and PAX5B gene products display differential expression signatures and tissue specificity[10][11].
3. PAX5 Expression and Regulation
As depicted in Figure 13, PAX5 is widely associated with various cellular processes and pathologies (Figure 3). Given the essential role of PAX5-mediated transactivation of vital genes for cell biology, its deregulation will have consequences on basic cellular processes such as differentiation, viability, and proliferation (reviewed in[4][12][13] ). Investigation of deregulated mechanisms leading to aberrant PAX5 expression and activity is therefore relevant and warranted to provide more insight into the overall comprehension of PAX5 mechanisms of action. Although the literature provides abundant research characterizing PAX5-mediated pathways and interactions, the upstream mechanisms regulating PAX5 expression are much less defined.
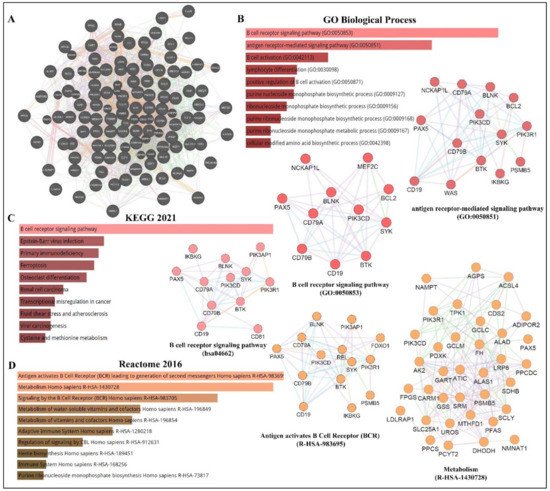
Figure 13. PAX5 interaction networks and related biological pathways. (A) PAX5 gene interaction networks have been mapped using the Cytoscape plugin GeneMANIA (https://genemania.org, accessed on 8 April 2022). Schematic illustrations of functional annotations and biological terms visualization are represented by: (B) PAX5 gene ontology (GO) in terms of functional orthologs and their relative implication in each predicted biological processes; (C) PAX5 pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, which provides an integrated evaluation of genomic, chemical, and biochemical functions; and (D) relative functional association to biological reactomes based on PAX5-related network genes. Annotations were done using the Enrichr algorithms (https://maayanlab.cloud/Enrichr, accessed on 10 April 2022). Significance was considered if p < 0.05.
4.
PAX5
Epigenetic Regulation
Many genomic studies have described the PAX5 locus as a genetic hot-spot susceptible to structural variation[14][15][16][17][18]. For example, PAX5 expression and function are altered by various genetic alterations, including somatic mutation, translocation, and duplication/polyploidy[19][8][20][21][22]. In addition to genetic mutation, which changes both the transcriptional levels and protein sequences, genes are also submitted to epigenetic deregulation, which impacts overall expression levels[23]. These epigenetic processes include methylation of 5′-cytosine-phosphate-guanine-3′ (CpG) islands, chromatin remodeling via histone modifications, and various RNA-mediated mechanisms, which involve regulatory non-coding RNAs[23][24]. A brief description of each regulatory mechanism and its impact on PAX5-mediated function is discussed below.
First, methylation of CpG islands to form 5′-methylcytosine (5mC) is a well-described mechanism to repress transcriptional expression of unwanted genes during fundamental cellular processes such as development and differentiation[25][26][27]. DNA methylation is catalyzed by a group of DNA methyltransferase (DNMT) enzyme members (e.g., DNMT1, DNMT3a, and DNMT3b)[28][29]. DNA methylation can also be reversed by demethylation, which is mediated by Ten-Eleven Translocation (TET) family dioxygenase enzymes, which include TET1, TET2, and TET3[30]. In fact, B-lineage development is coordinated by the well-timed deployment of B-cell fate transcription factors, which are regulated by epigenetic events and post-transcriptional modifications[31][32]. For example, DNMT1, DNMT3a, and DNMT3b are required for the maturation of hematopoietic stem cells into CLPs, whereas DNMT1 is particularly essential for pre-B-cell differentiation to immature B-cell[26][33]. Subsequent studies have since demonstrated that TET function is required for developing B-cells to transit from the pro-B to pre-B developmental stage[34]. Mechanistically, the B-cell-specific MB-1 (CD79a) promoter is known to be hypermethylated during hematopoietic stem cells transition to CLPs and then progressively demethylated during the expression and assembling of the BCR components. These events upregulate PAX5 expression and concomitant target genes to achieve B-lineage identity[35][36][37][38]. On the other hand, attenuation of PAX5 expression during terminal B-cell differentiation is reported to partly mediated by methylation of PAX5[39]. In support of these events, a study by Danbara et al., (2002) demonstrates that genomic demethylation using 5-aza-2′-deoxycytidine in myeloma cell lines results in the reconstitution of PAX5 expression and its transcriptional target genes (CD19 and MB-1)[39]. Although the regulation of the complex networks of epigenetic modifications governing B-cell differentiation is only partially understood, one aberrant mechanism leading to deregulated PAX5 methylation has been described for the inadequate function of AID[40][41]. The PAX5/AID pathway is essential for somatic hypermutation and antibody class switching during Ig production[42]. However, constitutive expression of AID has been associated with lymphomagenesis through its capacity to alter the sequence of non-Ig genes (i.e., PAX5) or through AID-mediated deamination of the PAX5 gene[41][43]. As a result, changes in PAX5 gene sequences redefine motif-specific regions marked for epigenetic modifications and subsequent expression control[40][41].
Given the importance of adequate methylation processes regulating PAX5-induced B-cell development, deregulated methylation results in the destabilization of B-cell homeostasis and cancer phenotypes[38][44]. This phenomenon has been further substantiated by the demonstration that PAX5 methylation status directly correlates with overall survival rates of cancer patients[45][46][47]. Furthermore, studies profiling methylation signatures in pediatric ALL patients have correlated PAX5 hypermethylation to the pathogenesis of B-ALL and T-ALL subtypes[48][49]. These findings have also prompted Nordlund et al. (2015) to propose that PAX5 methylation status combined with the mapping of PAX5 gene recombinations with other partner genes represent an effective diagnostic tool to classify heterogeneous and cytogenetically undefined ALL subtypes[50].
PAX5 aberrant methylation is not a tissue-specific phenomenon. In fact, PAX5 hypermethylation has been described in many non-hematological cancers, particularly where PAX5 is characterized as a tumor suppressor (e.g., hepatocellular carcinoma[51], ovarian carcinoma[51]; head and neck cancer[52], gastric cancer[47], lung and breast cancer malignancies[53][53]. Mechanistically, many of these latter studies demonstrate that silencing of PAX5 expression by hypermethylation leads to the inadequate transactivation of Tp53 expression, thus ensuing uncontrolled proliferation or decreased chemosensitivity to anticancer treatment regimens[45][54][55].
Gene expression profiles are also epigenetically regulated by multiple histone-modifying enzymes, which change chromatin structure to alter promoter region accessibility and recruit other modifications[56]. Histones, which assemble the nucleosomes, are prone to modifications, which include acetylation, methylation, ubiquitination, phosphorylation, and sumoylation[56]. The most common modifications consist of arginine methylation and/or lysine acetylation, where acetylation generally promotes gene expression whereas methylation elicits the opposite effects. Many histone modifying enzymes have been characterized including histone acetyltransferases (HATs), histone deacetylases (HDACs), histone demethylases, and various methyltransferases (e.g., Euchromatic Histone-Lysine N-Methyltransferase-2/EHMT2 and Lysine Methyltransferase-2A[26][44]. Like CpG island methylation, chromatin modifications represent an intrinsic part of B-cell activation and differentiation. For example, during early B-cell development, PAX5 secures B-cell commitment through activating B-cell specific genes. In addition, PAX5 concomitantly inhibits B-lineage inappropriate genes through the recruitment of HDACs to modify and silence promoter activation of these genes[38]. Studies show that the PAX5 locus is also continuously regulated by histone modifications throughout B-cell maturation. Specifically, the PAX5 promoter in pro-B-cells are modified by HDACs, whereas EHMT2 regulates mature B-cells located in germinal centers[26]. Another example is the EBF transcription factor, which is shown to be implicated in PAX5 and CD19 transactivation through the silencing of Lysine Methyltransferase-2A during early B-cell development[32][57][58]. Another example is the previously mentioned PAX5/BLIMP-1 axis during terminal B-cell differentiation into plasma cells. It is reported that BLIMP-1 suppresses PAX5 expression through the recruitment of histone demethylases and EHMT2 activities on the PAX5 promoter[59][60]. Furthermore, transcription factor Forkhead Box Protein-O1, which is essential for B-cell development beyond the pro-B-cell stage[61], is only activated upon histone methylation of TCF3, which only then can elicit histone modifications and silence PAX5 to enable the progression of B-cell development[62]. Another study conducted by Danbara et al., (2002) has specifically demonstrated that the upstream PAX5 promoter (exon 1A) is predominantly inactivated by DNA methylation, whereas the downstream promoter (exon 1B) is repressed by histone deacetylation during the final stages of B-cell terminal differentiation[39]. Comprehensively, deregulation of histone modifying events on PAX5 or its upstream regulators lead to aberrant PAX5 transcript levels and the development of diseases[44]. Accordingly, a recent study from Jin et al. (2021) has not only shown that PAX5 is hypermethylated in retinoblastoma tumors but also, the treatment of patients with cyclophosphamide (a common antineoplastic agent to treat retinoblastoma) increases PAX5 expression via gene demethylation and concomitant DNMT inhibition, which result in tumor regression[63][64].
To add complexity and appreciation for epigenetic mechanisms, different ATP-dependent chromatin remodeling complexes (CRC) capable of moving, ejecting, or restructuring nucleosomes (events often associated with DNA repair) have also been associated with PAX5 regulation and function[37][65]. For example, SWItch/Sucrose Non-Fermentable and the Nucleosome Remodeling Deacetylase CRCs are known to mediate PAX5-dependant induction or repression respectively of MB-1 (CD79a) gene expression during BCR assembly[37]. Therefore, the opposing functions of CRCs provide another layer of PAX5 function during B-cell development[37]. Another example is the histone modifying enzymes HATs, which can acetylate other cellular proteins (e.g., transcription factors) besides histones. A study by He et al., (2011) has found that histone acetyltransferase E1A binding protein p300 interacts with the C-terminal region of PAX5 to acetylate multiple lysine residues of the paired box DNA binding domain[66]. They also demonstrate that acetylation of the PAX5 transcription factor dramatically enhances the transactivation potential of its target genes[66]. This interaction was also investigated in B-cell lymphoma, where the Metastasis-Associated Protein-1 represents a substrate for acetylation upon its interaction with the HAT p300[67]. This study found that Metastasis-Associated Protein-1 acetylation leads to the direct transactivation and overexpression of PAX5, a widespread phenomenon in human DLBCL[67].
The final contributing mechanism in epigenetic control is mediated by non-coding RNAs, which include small interfering RNAs, microRNAs (miRNAs), piwi-interacting RNAs, long non-coding RNAs, and circular RNAs (circRNAs)[68][69][70][71]. In comparison to DNA and histone modifications, only a paucity of studies has directly elucidated ncRNA-mediated mechanisms governing PAX5 expression and function. A recent study from Harquail et al., (2019) has used a bioinformatic approach to establish a causal link between differentially expressed miRNAs in cancer cells in relation to their putative targeting of PAX5-dependent cancer processes and identified miRs-484 and 210 as directly regulators for PAX5 expression and function[72]. Interestingly, miR-210 has been extensively studied as a potent oncogenic miRNA, which targets critical tumor suppressors such as E2F3 and Tp53[73][74]. It is also well established that miR-210 is upregulated during hypoxia to induce EMT and tumor progression[75][76][77]. Given the prevalent role of PAX5 in epithelialization and EMT-MET processes in breast cancer cells[78][79], it has been suggested that miR-210 likely targets PAX5 during tumor neoplasm and hypoxia to produce a robust, comprehensive shift from epithelial to mesenchymal phenotypic features to evade hypoxic insult[72]. PAX5 has also been reported to be part of a regulatory feedback loop with miR-155 in cancer cells[80]. MiR-155 is known to play a vital role in the differentiation of memory B-cells where it targets PU.1 and AID necessary for B-cell commitment into plasma cell[81][82]. Despite the rapidly growing field of non-coding RNA function in biological processes, the elucidation of non-coding RNA-dependent control of PAX5 expression and function in B-cell development and disease is still under investigation. As our knowledge expands on the deregulation of miRNA profiles and its impact on biological processes, researchers notice that changes to the mRNA sequences targeted by miRNAs will also have significant consequences, including miRNA motif accessibility and disruption of translational control. Accordingly, the next section will discuss PAX5 post-transcriptional modifications and editing, which alter miRNA-specific targeting and impede the potential binding capacity of any motif-specific interacting partners of PAX5 products.
5. PAX5 Post-Transcriptional Regulation
Similar to most human gene transcripts, PAX5 mRNAs undergo alternative splicing processes, which translate into altered translational reading frames and often multiple protein isoforms[83][84][85][86]. To date, alternative splicing events of PAX5 transcripts in humans and other species result in translated products with deleted regions corresponding to single or multiple coding exons[83][85][86]. Specifically, studies have shown that alternative splicing of the 5′ or 3′ end of PAX5 mRNA leads to structural and functional alterations of the PAX5 transcription factor in the DNA binding (exons 2-3) and transactivation domains (exons 8-9) respectively[85][87][88]. A study performed by Robichaud et al., (2004) has characterized alternatively spliced PAX5 transcripts in CD19+ peripheral blood lymphocytes from healthy adult donors and found that B-cells simultaneously co-express multiple isoforms, including full-length mRNA (exons 1-10), in addition to transcripts lacking either exon 7 (∆7); exon 8 (∆8); exon 9 (∆9); exons 7-8 (∆7/8); or exons 7-8-9 (∆7/8/9)[85]. Interestingly, this study also demonstrates that each PAX5 protein variant elicits a unique transactivation potential upon downstream target genes[85]. Other studies have since reported additional C-terminal isoforms lacking exons 6-7-8-9 (∆6/7/8/9); exons 6-7-8 (∆6/7/8); exons 8-9 (∆8/9); and finally, a transcript containing a partial intronic sequence (intron 6) in healthy B-cells and lymphoma[88]. These findings underscore the complexity of potential dominant-negative effects and the outcome of downstream target gene expression due to a network of multiple PAX5 transcription factor variants. Despite various reports characterizing the expression of alternatively spliced PAX5 variants, the specific role of each isoform and their capacity to compete for putative PAX5 targets are still undefined. However, one study conducted by Sadakane et al., (2007) has correlated a specific expression profile comprising of the wild-type and the ∆8 PAX5 variants in over 90% of childhood acute lymphoblastic leukemia samples tested[89]. These findings suggest a possible role for individual PAX5 alternatively spliced isoforms in the regulation (or deregulation) of PAX5 function.
More recently, PAX5 transcripts have also been characterized to undergo 3′end shortening[90]. This type of transcriptional modification has significant repercussions on translational fate given that mRNA untranslated regions (UTRs), notably at the 3′end, harbor multiple binding sites for RNA binding proteins and other translational regulatory elements (e.g., miRNAs), which control transcript stability and translation efficiency[91][92]. A study by Beauregard et al., (2021) has recently reported that although 3′-editing of PAX5 transcripts is prevalent in healthy peripheral B-cells, shortening of the 3′UTR is directly linked to increased translation of PAX5 and correlates with leukemic disease progression[93]. Mechanistically, the study reveals that PAX5 3′UTR shortening is mainly due to sequence excision (up to 86%) by alternative splicing events. PAX5 mRNA shortening was also investigated in non-hematological cancers. Interestingly, conversely to 3′UTR splicing in B-cells, PAX5 3′UTR shortening in breast cancer cells is primarily manifested by alternative polyadenylation (APA)[94]. APA is another type of post-transcriptional modification where gene transcription is prompted to use alternative polyadenylation motifs (transcription termination signals), which alter the overall length of the mRNA sequences at their 3′ end. In fact, APA motifs are prevalent in more than half of all human transcripts, notably in oncogenes, to evade translational control at their 3′UTR, resulting in increased mRNA stability and translation[95][96][97]. To further elucidate the impact of PAX5 3′UTR shortening on miRNA targeting and regulation, a bioinformatic approach was used to identify predicted miRNAs targeting the excised 3′UTR in truncated PAX5 transcripts from cancer cells[98]. The study then experimentally validated that miR-181a, miR-217, and miR-1275 represent the most impactful tumor suppressors lost during PAX5 3′UTR shortening in cancer cell models[99]. Nevertheless, more studies are required to fully understand the impact of regulatory elements (e.g., miRNAs) and the accessibility of their corresponding binding sites deleted from truncated PAX5 transcripts in oncogenic processes and disease.
The previous sections describe multiple regulatory mechanisms, which lead to very diverse PAX5 transcripts, proteins, and functions. More recently, researchers have also characterized a new class of transcriptional PAX5 products, circular PAX5 RNAs (circPAX5)[100][101][102]. Circular RNAs (circRNAs) represent a relatively new category of non-coding RNAs characterized by a covalent phosphodiester bond between the 5′ and 3′ extremities of the transcript[103][104]. After being discovered in viruses in 1976, circular RNA was first observed in humans in 1991, when it was initially thought to be the product of improper post-translational editing[105]. Since then, circRNAs have been shown to be abundantly expressed and play essential roles in cell biology and disease[71][103]. Accordingly, circular RNAs can encode proteins through cap-independent translation pathways, regulate gene transcription, regulate gene translation, interact with proteins, and even mop up (sponge) small RNAs such as miRNAs[106][107][108]. Due to their essential role in cellular processes, circular RNA aberrant expression and function are consequently associated with diseases, including cancer[105][109][110]. In fact, a study by Gaffo et al. (2019) describes circPAX5 as one of the most differentially overexpressed products in pediatric B-ALL patients[100]. They also demonstrate that circPAX5 directly binds to miR-124-5p in B-cell precursors to promote B-ALL progression through the interference of the B-cell maturation process[100]. More recently, researchers have mapped multiple circPAX5 isoforms in B-cells including: circPAX5_2-3 (containing exons 2 and 3); circPAX5_2-4 (exons 2, 3 and 4); circPAX5_2-5 (exons 2, 3, 4 and 5); circPAX5_2-6 (exons 2, 3, 4, 5 and 6); circPAX5_2-7 (exons 2, 3, 4, 5, 6, and 7); circPAX5_2-8 (exons 2, 3, 4, 5, 6, 7, and 8); circPAX5_8 (exon 8); circPAX5_7-8 (exons 7 and 8); circPAX5_5-8 (exons 5, 6, 7, 8); and finally, circPAX5_2-6+intron 5 (exons 2, 3, 4, 5, partial intron 5, and exon 6)[111][112]. Furthermore, using TaqMan probes designed to target each unique circPAX5 junction region created by both extremities, researchers demonstrate that circPAX5_2-5 and circPAX5_2-6 are overexpressed in chronic lymphocytic leukemia patients in comparison to peripheral B-cells from healthy individuals. Mechanistically, researchers demonstrate that circPAX5 products interact with important microRNAs such as miR-146a and the miR-17-92 cluster. Previous reports demonstrate that miR-146a is a critical regulator of BLIMP-1 during B-cell differentiation[113], whereas microRNAs from the miR-17-92 cluster mediate the developmental transition of pro-B to pre-B-cells[114][115]. In addition, the miR-17-92 cluster is also associated with many oncogenic processes and phenotypes of hematopoietic cancers, notably in Burkitt lymphoma[116]. Altogether, these findings not only identify a new class of PAX5 products (i.e., circPAX5) but also provide new potential signaling avenues for PAX5-mediated function in B-cell development and disease.
6. Post-Translational Regulation of PAX5
Post-translational modifications and regulation of PAX5 function have not been extensively characterized. However, a few studies have reported specific PAX5-interacting regulators, which modify the PAX5 transcription factor to regulate its transactivation potential. As described earlier, PAX5 can be acetylated by HATs on multiple lysine residues, which enhances its transcriptional activation of downstream target genes[66]. Another example is how the PAX5 transcription factor can be regulated through phosphorylation events. Accordingly, studies show that PAX5 phosphorylation is responsible for the BLIMP-1/PAX5 regulatory axis during the critical stages of plasma cell differentiation[117][118]. Specifically, upon BCR engagement of pro-B cells, PAX5 is phosphorylated on serine and tyrosine residues by Extracellular Regulated Kinases-1/2 and Spleen Associated Tyrosine Kinase respectively, which revoke PAX5′s ability to repress BLIMP-1 expression, thus enabling the progression of plasma cell development. On the other hand, a study conducted by Kovac et al., (2000) has demonstrated that Importin alpha-1 interacts with the nuclear localization signal on PAX5 to confer its nuclear localization and import, leading to greater PAX5 transactivation of downstream target genes[119].
7. Discussion
It is well established that PAX5 products are important regulators of cell biology, notably in B-lineage commitment and maturation. It is also apparent that PAX5 is plagued not only by the high-profile genes it regulates but also, by its incredible vulnerability to genetic alterations leading to aberrant expression of PAX5 products. Given the reliance of crucial developmental program genes on PAX5 transactivity, perturbation of PAX5 expression and function at any level ultimately derails basic cellular processes, lending way to oncogenic manifestations. Moreover, given the requirements for coordinated and transitional PAX5 expression profiles during early (PAX5 activation) and late (PAX5 attenuation) phases of B-cell maturation, inadequate PAX5 activity leads to blockade of B-cell differentiation and uncontrolled proliferation of immature B-cells [7,62–64].
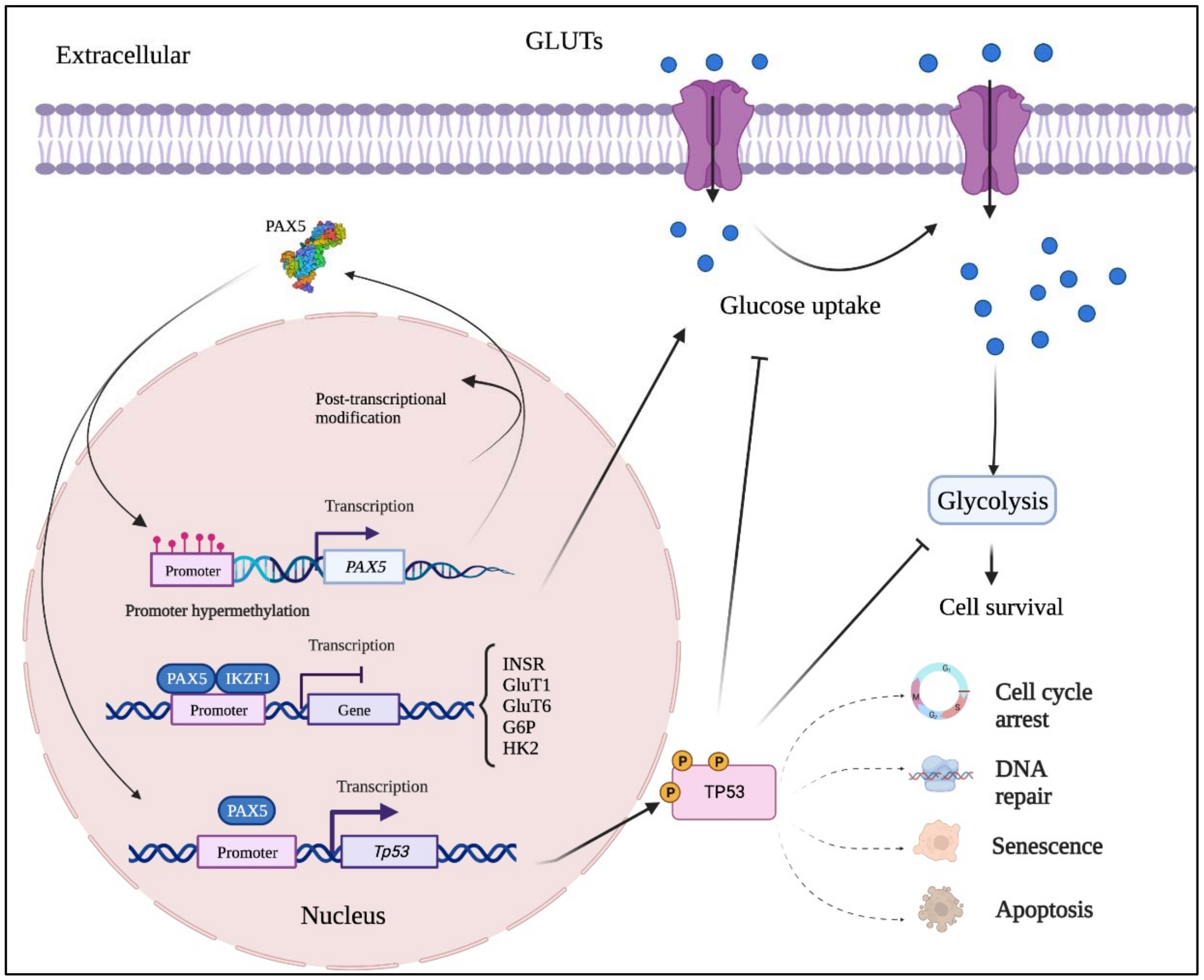

Figure 24. Mutation-independent mechanisms leading to aberrant PAX5 signaling and cell processes. Aside from PAX5 gene sequence alterations, deregulated PAX5 expression can also result from epigenetic events and post-transcription modifications. First, PAX5 gene promoter hypermethylation has been described in many cancers, notably when PAX5 behaves as a tumor suppressor. Post- transcriptional modifications (e.g., coding exon alternative splicing, 3′UTR shortening, and RNA circularization) also contribute to overall PAX5 expression and function. The net production of functional PAX5 transcription factors can thereafter collaborate with IKZF1 to regulate downstream metabolic genes to limit glucose uptake and energy supply required for oncogenic transformation. Adequate PAX5 function is also required to regulate Tp53 expression and avoid uncontrolled cancer phenotypes. Tp53 is also intimately linked to metabolic disfunction leading to cancer processes.
References
- Brian Thompson; Emily A. Davidson; Wei Liu; Daniel W. Nebert; Elspeth A. Bruford; Hongyu Zhao; Emmanouil T. Dermitzakis; David C. Thompson; Vasilis Vasiliou; Overview of PAX gene family: analysis of human tissue-specific variant expression and involvement in human disease. Quality of Life Research 2020, 140, 381-400, 10.1007/s00439-020-02212-9.
- Qiuyu Wang; Wen-Hui Fang; Jerzy Krupinski; Shant Kumar; Mark Slevin; Patricia Kumar; Paxgenes in embryogenesis and oncogenesis. Journal of Cellular and Molecular Medicine 2008, 12, 2281-2294, 10.1111/j.1582-4934.2008.00427.x.
- Darrell Underhill; PAX Proteins and Fables of Their Reconstruction. Critical Reviews? in Eukaryotic Gene Expression 2011, 22, 161-177, 10.1615/critreveukargeneexpr.v22.i2.70.
- Ewan J. D. Robson; Shu-Jie He; Michael R. Eccles; A PANorama of PAX genes in cancer and development. Nature Cancer 2005, 6, 52-62, 10.1038/nrc1778.
- D. Alan Underhill; Genetic and biochemical diversity in the Pax gene family. Biochemistry and Cell Biology 1999, 78, 629-638, 10.1139/bcb-78-5-629.
- B Adams; P Dörfler; A Aguzzi; Z Kozmik; P Urbánek; I Maurer-Fogy; M Busslinger; Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis.. Genes & Development 1992, 6, 1589-1607, 10.1101/gad.6.9.1589.
- Joris Andrieux; Sandra Fert-Ferrer; Marie-Christine Copin; Pauline Huyghe; Pierre Pocachard; James Lespinasse; Francis Bauters; Jean Luc Laï; Bruno Quesnel; Three new cases of non-Hodgkin lymphoma with t(9;14)(p13;q32). Cancer Genetics and Cytogenetics 2003, 145, 65-69, 10.1016/s0165-4608(03)00054-2.
- M Busslinger; N Klix; P Pfeffer; P G Graninger; Z Kozmik; Deregulation of PAX-5 by translocation of the Emu enhancer of the IgH locus adjacent to two alternative PAX-5 promoters in a diffuse large-cell lymphoma.. Proceedings of the National Academy of Sciences 1996, 93, 6129-6134, 10.1073/pnas.93.12.6129.
- A Barberis; K Widenhorn; L Vitelli; M Busslinger; A novel B-cell lineage-specific transcription factor present at early but not late stages of differentiation.. Genes & Development 1990, 4, 849-859, 10.1101/gad.4.5.849.
- Charlotte Cresson; Sophie Péron; Laura Jamrog; Nelly Rouquié; Nais Prade; Marine Dubois; Sylvie Hébrard; Stéphanie Lagarde; Bastien Gerby; Stéphane J.C. Mancini; et al.Michel CognéEric DelabesseLaurent DelpyCyril Broccardo PAX5A and PAX5B isoforms are both efficient to drive B cell differentiation. Oncotarget 2018, 9, 32841-32854, 10.18632/oncotarget.26003.
- Hidehiko Kikuchi; Masami Nakayama; Futoshi Kuribayashi; Hitomi Mimuro; Hideki Nishitoh; Yasunari Takami; Tatsuo Nakayama; Shinobu Imajoh-Ohmi; Paired box gene 5 isoforms A and B have different functions in transcriptional regulation of B cell development-related genes in immature B cells. Microbiology and Immunology 2015, 59, 426-431, 10.1111/1348-0421.12272.
- Mohammad Shahjahani; Fatemeh Norozi; A. Ahmadzadeh; Saeid Shahrabi; Farzaneh Tavakoli; Ali Aminasnafi; Najmaldin Saki; The role of Pax5 in leukemia: diagnosis and prognosis significance. Medical Oncology 2014, 32, 1-8, 10.1007/s12032-014-0360-6.
- Pierre O'Brien; Pier Morin; Rodney J. Ouellette; Gilles A. Robichaud; The Pax-5 Gene: A Pluripotent Regulator of B-cell Differentiation and Cancer Disease. Cancer Research 2011, 71, 7345-7350, 10.1158/0008-5472.can-11-1874.
- Sarah E. Arthur; Aixiang Jiang; Bruno M. Grande; Miguel Alcaide; Razvan Cojocaru; Christopher K. Rushton; Anja Mottok; Laura K. Hilton; Prince Kumar Lat; Eric Y. Zhao; et al.Luka CulibrkDaisuke EnnishiSelin JessaLauren ChongNicole ThomasPrasath PararajalingamBarbara MeissnerMerrill BoyleJordan DavidsonKevin R. BushellDaniel LaiPedro FarinhaGraham W. SlackGregg B. MorinSohrab ShahDipankar SenSteven J. M. JonesAndrew J. MungallRandy D. GascoyneTimothy E. AudasPeter UnrauMarco A. MarraJoseph M. ConnorsChristian SteidlDavid W. ScottRyan D. Morin Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nature Communications 2018, 9, 4001, 10.1038/s41467-018-06354-3.
- Xose S. Puente; Silvia Beà; Rafael Valdés-Mas; Neus Villamor; Jesús Gutiérrez-Abril; José I. Martín-Subero; Marta Munar; Carlota Rubio-Pérez; Pedro Jares; Marta Aymerich; et al.Tycho BaumannRenée BeekmanLaura BelverAnna CarrioGiancarlo CastellanoGuillem ClotEnrique ColadoDolors ColomerDolors CostaJulio DelgadoAnna EnjuanesXavier EstivillAdolfo A. FerrandoJosep L. GelpíBlanca GonzálezSantiago GonzálezMarcos GonzálezMarta GutJesús M. Hernández-RivasMónica López-GuerraDavid Martín-GarcíaAlba NavarroPilar NicolásModesto OrozcoÁngel R. PayerMagda PinyolDavid G. PisanoDiana A. PuenteAna C. QueirósVíctor QuesadaCarlos M. Romeo-CasabonaCristina RoyoRomina RoyoMaría RozmanNuria RussiñolItziar SalaverríaKostas StamatopoulosHendrik G. StunnenbergDavid TamboreroMaría J. TerolAlfonso ValenciaNuria López-BigasDavid TorrentsIvo GutArmando López-GuillermoCarlos López-OtínElías Campo Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015, 526, 519-524, 10.1038/nature14666.
- Bruno M. Grande; Daniela S. Gerhard; Aixiang Jiang; Nicholas B. Griner; Jeremy S. Abramson; Thomas B. Alexander; Hilary Allen; Leona W. Ayers; Jeffrey M. Bethony; Kishor Bhatia; et al.Jay BowenCorey CasperJohn Kim ChoiLuka CulibrkTanja M. DavidsenMaureen A. DyerJulie M. Gastier-FosterPatee GesuwanTimothy C. GreinerThomas G. GrossBenjamin HanfNancy Lee HarrisYiwen HeJohn D. IrvinElaine S. JaffeSteven J. M. JonesPatrick KerchanNicole KnoetzeFabio E. LealTara M. LichtenbergYussanne MaJean Paul MartinMarie-Reine MartinSam M. MbulaiteyeCharles G. MullighanAndrew J. MungallConstance NamirembeKaren NovikAriela NoyMartin D. OgwangAbraham OmodingJackson OremSteven J. ReynoldsChristopher K. RushtonJohn T. SandlundRoland SchmitzCynthia TaylorWyndham H. WilsonGeorge W. WrightEric Y. ZhaoMarco A. MarraRyan D. MorinLouis M. Staudt Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 2019, 133, 1313-1324, 10.1182/blood-2018-09-871418.
- Merja Heinäniemi; Tapio Vuorenmaa; Susanna Teppo; Minna U Kaikkonen; Maria Bouvy-Liivrand; Juha Mehtonen; Henri Niskanen; Vasilios Zachariadis; Saara Laukkanen; Thomas Liuksiala; et al.Kaisa TeittinenOlli Lohi Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots. eLife 2016, 5, -, 10.7554/elife.13087.
- Marta Kasprzyk; Weronika Sura; Agnieszka Dzikiewicz-Krawczyk; Enhancing B-Cell Malignancies—On Repurposing Enhancer Activity towards Cancer. Cancers 2021, 13, 3270, 10.3390/cancers13133270.
- N Kawamata; M A Pennella; J L Woo; A J Berk; H P Koeffler; Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene 2011, 31, 966-977, 10.1038/onc.2011.291.
- J Familiades; Marina Bousquet; M Lafage-Pochitaloff; M-C Béné; K Beldjord; John De Vos; N Dastugue; E Coyaud; S Struski; C Quelen; et al.N Prade-HoudellierS DobbelsteinJ-M CayuelaJ SoulierN GrardelC PreudhommeHélène CavéO BlanchetV LhéritierA DelannoyY ChalandonN IfrahA PigneuxP BroussetE A MacintyreF HuguetH DombretC BroccardoE DelabesseM-C B[Eacute]N PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia 2009, 23, 1989-1998, 10.1038/leu.2009.135.
- Sohela Shah; Kasmintan Schrader; Esmé Waanders; Andrew E. Timms; Joseph Vijai; Cornelius Miething; Jeremy Wechsler; Jun Yang; James E Hayes; Robert Klein; et al.Jinghui ZhangLei WeiGang WuMichael RuschPanduka NagahawatteJing MaShann-Ching ChenGuangchun SongJinjun ChengPaul MeyersDeepa BhojwaniSuresh C JhanwarPeter MaslakMartin FleisherJason LittmanLily OffitRohini Rau-MurthyMegan Harlan FleischutMarina CorinesRajmohan MuraliXiaoni GaoChristopher ManschreckThomas KitzingVundavalli V. MurtySusana C RaimondiRoland P. KuiperAnnet SimonsJoshua D. SchiffmanKenan OnelSharon E. PlonDavid A WheelerDeborah RitterDavid ZieglerKatherine L TuckerRosemary SuttonGeorgia Chenevix-TrenchJun LiDavid G. HuntsmanSamantha HansfordJanine SenzTom WalshMing LeeChris HahnKathryn G RobertsMary-Claire KingSarah M. LoRoss L. LevineAgnes VialeNicholas D. SocciKatherine NathansonHamish S. ScottMark J DalySteven M. LipkinScott W. LoweJames R. DowningDavid AltshulerJohn T. SandlundMarshall S. HorwitzCharles G. MullighanKenneth Offit A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nature Genetics 2013, 45, 1226-1231, 10.1038/ng.2754.
- Etienne Coyaud; Stephanie Struski; Nais Prade; Julien Familiades; Ruth Eichner; Cathy Quelen; Marina Bousquet; Francine Mugneret; Pascaline Talmant; Marie-Pierre Pages; et al.Christine LefebvreMinique PentherEric LippertNathalie NadalSylvie TaviauxBruce PoppeIsabelle LuquetLaurence BarangerVirginie EclacheIsabelle RadfordCarole BarinMarie-Joëlle MozziconacciMarina Lafage-PochitaloffHélène Antoine-PoirelChristiane CharrinChristine PerotChristine TerrePierre BroussetNicole DastugueCyril Broccardo Wide diversity of PAX5 alterations in B-ALL: a Groupe Francophone de Cytogénétique Hématologique study. Blood 2010, 115, 3089-3097, 10.1182/blood-2009-07-234229.
- E R Gibney; C M Nolan; Epigenetics and gene expression. Heredity 2010, 105, 4-13, 10.1038/hdy.2010.54.
- Jian-Wei Wei; Kai Huang; Chao Yang; Chun-Sheng Kang; Non-coding RNAs as regulators in epigenetics. Oncology Reports 2016, 37, 3-9, 10.3892/or.2016.5236.
- Barbara K. Dunn; Mukesh Verma; Asad Umar; Epigenetics in Cancer Prevention: Early Detection and Risk Assessment. Annals of the New York Academy of Sciences 2003, 983, 1-4, 10.1111/j.1749-6632.2003.tb05957.x.
- Yan Bao; Xuetao Cao; Epigenetic Control of B Cell Development and B-Cell-Related Immune Disorders. Clinical Reviews in Allergy & Immunology 2015, 50, 301-311, 10.1007/s12016-015-8494-7.
- Toshiaki Kurogi; Hiroko Inoue; Yun Guo; Asako Nobukiyo; Keiko Nohara; Masamoto Kanno; A Methyl-Deficient Diet Modifies Early B Cell Development. Pathobiology 2012, 79, 209-218, 10.1159/000337290.
- Mary Grace Goll; Timothy H. Bestor; EUKARYOTIC CYTOSINE METHYLTRANSFERASES. Annual Review of Biochemistry 2005, 74, 481-514, 10.1146/annurev.biochem.74.010904.153721.
- Renata Zofia Jurkowska; Tomasz Piotr Jurkowski; Albert Jeltsch; Structure and Function of Mammalian DNA Methyltransferases. ChemBioChem 2010, 12, 206-222, 10.1002/cbic.201000195.
- Fabian Mohr; Konstanze Döhner; Christian Buske; Vijay P.S. Rawat; TET Genes: new players in DNA demethylation and important determinants for stemness. Experimental Hematology 2011, 39, 272-281, 10.1016/j.exphem.2010.12.004.
- Guideng Li; Hong Zan; Zhenming Xu; Paolo Casali; Epigenetics of the antibody response. Trends in Immunology 2013, 34, 460-470, 10.1016/j.it.2013.03.006.
- Mohamed Amin Choukrallah; Patrick Matthias; The Interplay between Chromatin and Transcription Factor Networks during B Cell Development: Who Pulls the Trigger First?. Frontiers in Immunology 2014, 5, 156, 10.3389/fimmu.2014.00156.
- Sara R. Cherry; Caroline Beard; Rudolf Jaenisch; David Baltimore; V(D)J recombination is not activated by demethylation of the kappa locus. Proceedings of the National Academy of Sciences 2000, 97, 8467-8472, 10.1073/pnas.150218497.
- Chan-Wang Lio; Jiayuan Zhang; Edahí González-Avalos; Patrick G Hogan; Xing Chang; Anjana Rao; Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 2016, 5, e18290, 10.7554/elife.18290.
- Jasna Medvedovic; Anja Ebert; Hiromi Tagoh; Meinrad Busslinger; Pax5. null 2011, 111, 179-206, 10.1016/b978-0-12-385991-4.00005-2.
- Holly Maier; Rachel Ostraat; Hua Gao; Scott Fields; Susan A Shinton; Kay L Medina; Tomokatsu Ikawa; Cornelis Murre; Harinder Singh; Richard R Hardy; et al.James Hagman Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nature Immunology 2004, 5, 1069-1077, 10.1038/ni1119.
- Hua Gao; Kara Lukin; Julita Ramírez; Scott Fields; Desiree Lopez; James Hagman; Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proceedings of the National Academy of Sciences 2009, 106, 11258-11263, 10.1073/pnas.0809485106.
- Shane McManus; Anja Ebert; Giorgia Salvagiotto; Jasna Medvedovic; Qiong Sun; Ido Tamir; Markus Jaritz; Hiromi Tagoh; Meinrad Busslinger; The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. The EMBO Journal 2011, 30, 2388-2404, 10.1038/emboj.2011.140.
- Mikio Danbara; Kohzoh Kameyama; Masaaki Higashihara; Yohtaroh Takagaki; DNA methylation dominates transcriptional silencing of Pax5 in terminally differentiated B cell lines. Molecular Immunology 2002, 38, 1161-1166, 10.1016/s0161-5890(02)00003-2.
- Hiroyuki Gonda; Manabu Sugai; Yukiko Nambu; Tomoya Katakai; Yasutoshi Agata; Kazuhiro J. Mori; Yoshifumi Yokota; Akira Shimizu; The Balance Between Pax5 and Id2 Activities Is the Key to AID Gene Expression. Journal of Experimental Medicine 2003, 198, 1427-1437, 10.1084/jem.20030802.
- Pilar M Dominguez; Rita Eshaknovich; Epigenetic Function of Activation-Induced Cytidine Deaminase and Its Link to Lymphomagenesis. Frontiers in Immunology 2014, 5, 642, 10.3389/fimmu.2014.00642.
- Masamichi Muramatsu; Kazuo Kinoshita; Sidonia Fagarasan; Shuichi Yamada; Yoichi Shinkai; Tasuku Honjo; Class Switch Recombination and Hypermutation Require Activation-Induced Cytidine Deaminase (AID), a Potential RNA Editing Enzyme. Cell 2000, 102, 553-563, 10.1016/s0092-8674(00)00078-7.
- Laura Pasqualucci; Peter Neumeister; Tina Goossens; Gouri Nanjangud; R. S. K. Chaganti; Ralf Küppers; Riccardo Dalla-Favera; Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 2001, 412, 341-346, 10.1038/35085588.
- Haijing Wu; Yaxiong Deng; Yu Feng; Di Long; Kongyang Ma; Xiaohui Wang; Ming Zhao; Liwei Lu; Qianjin Lu; Epigenetic regulation in B-cell maturation and its dysregulation in autoimmunity. Cellular & Molecular Immunology 2018, 15, 676-684, 10.1038/cmi.2017.133.
- Keisuke Kurimoto; Masamichi Hayashi; Rafael Guerrero-Preston; Masahiko Koike; Mitsuro Kanda; Sho Hirabayashi; Hiroshi Tanabe; Nao Takano; Naoki Iwata; Yukiko Niwa; et al.Hideki TakamiDaisuke KobayashiChie TanakaSuguru YamadaGoro NakayamaHiroyuki SugimotoTsutomu FujiiMichitaka FujiwaraYasuhiro Kodera PAX5 gene as a novel methylation marker that predicts both clinical outcome and cisplatin sensitivity in esophageal squamous cell carcinoma. Epigenetics 2017, 12, 865-874, 10.1080/15592294.2017.1365207.
- X Li; K F Cheung; X Ma; L Tian; J Zhao; M Y Y Go; B Shen; A S L Cheng; J Ying; Q Tao; et al.J J Y SungH-F KungJ Yu Epigenetic inactivation of paired box gene 5, a novel tumor suppressor gene, through direct upregulation of p53 is associated with prognosis in gastric cancer patients. Oncogene 2011, 31, 3419-3430, 10.1038/onc.2011.511.
- Jingyu Deng; Han Liang; Rupeng Zhang; Qiuping Dong; Yachao Hou; Jun Yu; Daiming Fan; Xishan Hao; Applicability of the methylated CpG sites of paired box 5 (PAX5) promoter for prediction the prognosis of gastric cancer. Oncotarget 2014, 5, 7420-7430, 10.18632/oncotarget.1973.
- Gero Hütter; Martin Kaiser; Martin Neumann; Maximilian Mossner; Daniel Nowak; Claudia D. Baldus; Nicola Gökbuget; Dieter Hoelzer; Eckhard Thiel; Wolf-Karsten Hofmann; et al. Epigenetic regulation of PAX5 expression in acute T-cell lymphoblastic leukemia. Leukemia Research 2011, 35, 614-619, 10.1016/j.leukres.2010.11.015.
- Charles G. Mullighan; James R. Downing; Global Genomic Characterization of Acute Lymphoblastic Leukemia. Seminars in Hematology 2009, 46, 3-15, 10.1053/j.seminhematol.2008.09.005.
- Jessica Nordlund; Christofer L Bäcklin; Vasilios Zachariadis; Lucia Cavelier; Johan Dahlberg; Ingegerd Öfverholm; Gisela Barbany; Ann Nordgren; Elin Övernäs; Jonas Abrahamsson; et al.Trond FlaegstadMats M HeymanÓlafur G JónssonJukka KanervaRolf LarssonJosefine PalleKjeld SchmiegelowMats G GustafssonGudmar LönnerholmErik ForestierAnn-Christine Syvänen DNA methylation-based subtype prediction for pediatric acute lymphoblastic leukemia. Clinical Epigenetics 2015, 7, 11-11, 10.1186/s13148-014-0039-z.
- Martin Mžik; Marcela Chmelařová; Stanislav John; Jan Laco; Ondřej Slabý; Igor Kiss; Lucia Bohovicová; Vladimir Palicka; Jana Nekvindová; Aberrant methylation of tumour suppressor genes WT1, GATA5 and PAX5 in hepatocellular carcinoma. Clinical Chemistry and Laboratory Medicine (CCLM) 2015, 54, 1971-1980, 10.1515/cclm-2015-1198.
- Rafael Guerrero-Preston; Christina Michailidi; Luigi Marchionni; Curtis R Pickering; Mitchell J Frederick; Jeffrey N Myers; Srinivasan Yegnasubramanian; Tal Hadar; Maartje G Noordhuis; Veronika Zizkova; et al.Elana FertigNishant AgrawalWilliam WestraWayne KochJoseph CalifanoVictor E VelculescuDavid Sidransky Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics 2014, 9, 1031-1046, 10.4161/epi.29025.
- Cathy B Moelans; Anoek Hj Verschuur-Maes; Paul J van Diest; Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. The Journal of Pathology 2011, 225, 222-231, 10.1002/path.2930.
- Grace Guo; Guodong Li; Faculty Opinions recommendation of Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway.. Faculty Opinions – Post-Publication Peer Review of the Biomedical Literature 2011, 53, -, 10.3410/f.9126958.9711054.
- Weiwei Zhang; Wenji Yan; Niansong Qian; Quanli Han; Weitao Zhang; Guanghai Dai; Paired box 5 increases the chemosensitivity of esophageal squamous cell cancer cells by promoting p53 signaling activity. Chinese Medical Journal 2022, 135, 606-618, 10.1097/cm9.0000000000002018.
- Scott B. Rothbart; Brian D. Strahl; Interpreting the language of histone and DNA modifications. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2014, 1839, 627-643, 10.1016/j.bbagrm.2014.03.001.
- Ross Dickins; Grace Liu; Luisa Cimmino; Yifang Hu; Sarah Best; Laura Tuohey; Michael Farrar; Stephen Nutt; Gordon Smyth; PAX5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Experimental Hematology 2013, 41, S16, 10.1016/j.exphem.2013.05.059.
- Charles E. Bullerwell; Philippe Pierre Robichaud; Pierre M. L. Deprez; Andrew P. Joy; Gabriel Wajnberg; Darwin D’Souza; Simi Chacko; Sébastien Fournier; Nicolas Crapoulet; David A. Barnett; et al.Stephen M. LewisRodney J. Ouellette EBF1 drives hallmark B cell gene expression by enabling the interaction of PAX5 with the MLL H3K4 methyltransferase complex. Scientific Reports 2021, 11, 1-14, 10.1038/s41598-021-81000-5.
- Jin Yu; Cristina Angelin-Duclos; Jessica Greenwood; Jerry Liao; Kathryn Calame; Transcriptional Repression by Blimp-1 (PRDI-BF1) Involves Recruitment of Histone Deacetylase. Molecular and Cellular Biology 2000, 20, 2592-2603, 10.1128/mcb.20.7.2592-2603.2000.
- Ildikó Győry; Jian Wu; György Fejér; Edward Seto; Kenneth L Wright; PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nature Immunology 2004, 5, 299-308, 10.1038/ni1046.
- David Dominguez-Sola; Jennifer Kung; Antony B. Holmes; Victoria A. Wells; Tongwei Mo; Katia Basso; Riccardo Dalla-Favera; The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity 2015, 43, 1064-1074, 10.1016/j.immuni.2015.10.015.
- Yin Lin; Suchit Jhunjhunwala; Christopher Benner; Sven Heinz; Eva Welinder; Robert Mansson; Mikael Sigvardsson; James Hagman; Celso A Espinoza; Janusz Dutkowski; et al.Trey IdekerChristopher K GlassCornelis Murre A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nature Immunology 2010, 11, 635-643, 10.1038/ni.1891.
- Gabriella Livide; Maria Carmela Epistolato; Mariangela Amenduni; Vittoria Disciglio; Annabella Marozza; Maria Antonietta Mencarelli; Paolo Toti; Stefano Lazzi; Theodora Hadjistilianou; Sonia De Francesco; et al.Alfonso D’AmbrosioAlessandra RenieriFrancesca Ariani Epigenetic and Copy Number Variation Analysis in Retinoblastoma by MS-MLPA. Pathology and Oncology Research 2012, 18, 703-712, 10.1007/s12253-012-9498-8.
- Lan Jin; Xiaojie Ma; Xiaoqin Lei; Jing’An Tong; Runsheng Wang; Cyclophosphamide inhibits Pax5 methylation to regulate the growth of retinoblastoma via the Notch1 pathway. Human & Experimental Toxicology 2021, 40, S497-S508, 10.1177/09603271211051601.
- I. Whitehouse; A. Flaus; K. Havas; T. Owen-Hughes; Mechanisms for ATP-dependent chromatin remodelling. Biochemical Society Transactions 2000, 28, 376-379, 10.1042/0300-5127:0280376.
- Ti He; Sang Yong Hong; Lin Huang; Weihua Xue; Zhihong Yu; Hyoung Kwon; Marion Kirk; Shi-Jian Ding; Kaihong Su; Zhixin Zhang; et al. Histone Acetyltransferase p300 Acetylates Pax5 and Strongly Enhances Pax5-mediated Transcriptional Activity. Journal of Biological Chemistry 2011, 286, 14137-14145, 10.1074/jbc.m110.176289.
- Seetharaman Balasenthil; Anupama E. Gururaj; Amjad H. Talukder; Rozita Bagheri-Yarmand; Ty Arrington; Brian J. Haas; John C. Braisted; Insun Kim; Norman H. Lee; Rakesh Kumar; et al. Identification of Pax5 as a Target of MTA1 in B-Cell Lymphomas. Cancer Research 2007, 67, 7132-7138, 10.1158/0008-5472.can-07-0750.
- Daniel Holoch; Danesh Moazed; RNA-mediated epigenetic regulation of gene expression. Nature Reviews Genetics 2015, 16, 71-84, 10.1038/nrg3863.
- Anderson A Butler; William M Webb; Farah D Lubin; Regulatory RNAs and control of epigenetic mechanisms: expectations for cognition and cognitive dysfunction. Epigenomics 2015, 8, 135-151, 10.2217/epi.15.79.
- Hongyu Liu; Cheng Lei; Qin He; Zou Pan; Desheng Xiao; Yongguang Tao; Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Molecular Cancer 2018, 17, 1-14, 10.1186/s12943-018-0765-5.
- Lesca M. Holdt; Alexander Kohlmaier; Daniel Teupser; Molecular roles and function of circular RNAs in eukaryotic cells. Cellular and Molecular Life Sciences 2017, 75, 1071-1098, 10.1007/s00018-017-2688-5.
- Jason Harquail; Nicolas Leblanc; Rodney J Ouellette; Gilles A Robichaud; miRNAs 484 and 210 regulate Pax-5 expression and function in breast cancer cells. Carcinogenesis 2019, 40, 1010-1020, 10.1093/carcin/bgy191.
- Françoise Rothé; Michail Ignatiadis; Carole Chaboteaux; Benjamin Haibe-Kains; Naïma Kheddoumi; Samira Majjaj; Bassam Badran; Hussein Fayyad-Kazan; Christine Desmedt; Adrian Harris; et al.Martine PiccartChristos Sotiriou Global MicroRNA Expression Profiling Identifies MiR-210 Associated with Tumor Proliferation, Invasion and Poor Clinical Outcome in Breast Cancer. PLOS ONE 2011, 6, e20980, 10.1371/journal.pone.0020980.
- Xin Huang; Lianghao Ding; Kevin L. Bennewith; Ricky T. Tong; Scott Welford; K. Kian Ang; Michael Story; Quynh-Thu Le; Amato J. Giaccia; Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Molecular Cell 2009, 35, 856-867, 10.1016/j.molcel.2009.09.006.
- Qin Qin; Wei Furong; Li Baosheng; Multiple functions of hypoxia-regulated miR-210 in cancer. Journal of Experimental & Clinical Cancer Research 2014, 33, 50-50, 10.1186/1756-9966-33-50.
- Cecilia Devlin; Simona Greco; Fabio Martelli; Mircea Ivan; miR-210: More than a silent player in hypoxia. IUBMB Life 2011, 63, 94-100, 10.1002/iub.427.
- Xin Huang; Quynh-Thu Le; Amato J. Giaccia; MiR-210 – micromanager of the hypoxia pathway. Trends in Molecular Medicine 2010, 16, 230-237, 10.1016/j.molmed.2010.03.004.
- Laurent J-P Vidal; Jo K. Perry; Cecile M. Vouyovitch; Vijay Pandey; Severine E. Brunet-Dunand; Hichem C. Mertani; Dong-Xu Liu; Peter E. Lobie; PAX5α Enhances the Epithelial Behavior of Human Mammary Carcinoma Cells. Molecular Cancer Research 2010, 8, 444-456, 10.1158/1541-7786.mcr-09-0368.
- Sami Benzina; Annie-Pier Beauregard; Roxann Guerrette; Stéphanie Jean; Mame Daro Faye; Mark Laflamme; Emmanuel Maïcas; Nicolas Crapoulet; Rodney J. Ouellette; Gilles A. Robichaud; et al. Pax-5 is a potent regulator of E-cadherin and breast cancer malignant processes. Oncotarget 2017, 8, 12052-12066, 10.18632/oncotarget.14511.
- Jason Harquail; Nicolas LeBlanc; Carine Landry; Nicolas Crapoulet; Gilles A. Robichaud; Pax-5 Inhibits NF-κB Activity in Breast Cancer Cells Through IKKε and miRNA-155 Effectors. Journal of Mammary Gland Biology and Neoplasia 2018, 23, 177-187, 10.1007/s10911-018-9404-4.
- Dong Lu; Rinako Nakagawa; Sandra Lazzaro; Philipp Staudacher; Cei Abreu-Goodger; Tom Henley; Sara Boiani; Rebecca Leyland; Alison Galloway; Simon Andrews; et al.Geoffrey ButcherStephen NuttMartin TurnerElena Vigorito The miR-155–PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. Journal of Experimental Medicine 2014, 211, 2183-2198, 10.1084/jem.20140338.
- Kathryn Calame; MicroRNA-155 Function in B Cells. Immunity 2007, 27, 825-827, 10.1016/j.immuni.2007.11.010.
- Nancy D. Borson; Martha Q. Lacy; Peter J. Wettstein; Altered mRNA expression of Pax5 and Blimp-1 in B cells in multiple myeloma. Blood 2002, 100, 4629-4639, 10.1182/blood.v100.13.4629.
- Patty Zwollo; Hector Arrieta; Kaleo Ede; Karen Molinder; Stephen Desiderio; Roberta Pollock; The Pax-5 Gene Is Alternatively Spliced during B-cell Development. Journal of Biological Chemistry 1997, 272, 10160-10168, 10.1074/jbc.272.15.10160.
- Gilles A. Robichaud; Michel Nardini; Mark Laflamme; Miroslava Cuperlovic-Culf; Rodney J. Ouellette; Human Pax-5 C-terminal Isoforms Possess Distinct Transactivation Properties and Are Differentially Modulated in Normal and Malignant B Cells. Journal of Biological Chemistry 2004, 279, 49956-49963, 10.1074/jbc.m407171200.
- Elizabeth MacMurray; Maggie Barr; Amber Bruce; Lidia Epp; Patty Zwollo; Alternative splicing of the trout Pax5 gene and identification of novel B cell populations using Pax5 signatures. Developmental & Comparative Immunology 2013, 41, 270-281, 10.1016/j.dci.2013.06.008.
- Jillian Anspach; Gail Poulsen; Ilsa Kaattari; Roberta Pollock; Patty Zwollo; Reduction in DNA Binding Activity of the Transcription Factor Pax-5a in B Lymphocytes of Aged Mice. The Journal of Immunology 2001, 166, 2617-2626, 10.4049/jimmunol.166.4.2617.
- Jean-René Arseneau; Mark Laflamme; Stephen M. Lewis; Emmanuel Maïcas; Rodney J. Ouellette; Multiple isoforms ofPAX5are expressed in both lymphomas and normal B-cells. British Journal of Haematology 2009, 147, 328-338, 10.1111/j.1365-2141.2009.07859.x.
- Y. Sadakane; M. Zaitsu; M. Nishi; K. Sugita; S. Mizutani; A. Matsuzaki; E. Sueoka; Y. Hamasaki; E. Ishii; Expression and production of aberrant PAX5 with deletion of exon 8 in B-lineage acute lymphoblastic leukaemia of children. British Journal of Haematology 2006, 136, 297-300, 10.1111/j.1365-2141.2006.06425.x.
- Annie-Pier Beauregard; Brandon Hannay; Ehsan Gharib; Nicolas Crapoulet; Nicholas Finn; Roxann Guerrette; Amélie Ouellet; Gilles A. Robichaud; Pax-5 Protein Expression Is Regulated by Transcriptional 3′UTR Editing. Cells 2021, 11, 76, 10.3390/cells11010076.
- Catia Andreassi; Antonella Riccio; To localize or not to localize: mRNA fate is in 3′UTR ends. Trends in Cell Biology 2009, 19, 465-474, 10.1016/j.tcb.2009.06.001.
- A. Sachs; The role of poly(A) in the translation and stability of mRNA. Current Opinion in Cell Biology 1990, 2, 1092-1098, 10.1016/0955-0674(90)90161-7.
- Annie-Pier Beauregard; Brandon Hannay; Ehsan Gharib; Nicolas Crapoulet; Nicholas Finn; Roxann Guerrette; Amélie Ouellet; Gilles A. Robichaud; Pax-5 Protein Expression Is Regulated by Transcriptional 3′UTR Editing. Cells 2021, 11, 76, 10.3390/cells11010076.
- Annie-Pier Beauregard; Brandon Hannay; Ehsan Gharib; Nicolas Crapoulet; Nicholas Finn; Roxann Guerrette; Amélie Ouellet; Gilles A. Robichaud; Pax-5 Protein Expression Is Regulated by Transcriptional 3′UTR Editing. Cells 2021, 11, 76, 10.3390/cells11010076.
- Bin Tian; A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Research 2005, 33, 201-212, 10.1093/nar/gki158.
- Rickard Sandberg; Joel R. Neilson; Arup Sarma; Phillip A. Sharp; Christopher B. Burge; Proliferating Cells Express mRNAs with Shortened 3' Untranslated Regions and Fewer MicroRNA Target Sites. Science 2008, 320, 1643-1647, 10.1126/science.1155390.
- Christine Mayr; David P. Bartel; Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell 2009, 138, 673-684, 10.1016/j.cell.2009.06.016.
- Annie-Pier Beauregard; Brandon Hannay; Ehsan Gharib; Nicolas Crapoulet; Nicholas Finn; Roxann Guerrette; Amélie Ouellet; Gilles A. Robichaud; Pax-5 Protein Expression Is Regulated by Transcriptional 3′UTR Editing. Cells 2021, 11, 76, 10.3390/cells11010076.
- Annie-Pier Beauregard; Brandon Hannay; Ehsan Gharib; Nicolas Crapoulet; Nicholas Finn; Roxann Guerrette; Amélie Ouellet; Gilles A. Robichaud; Pax-5 Protein Expression Is Regulated by Transcriptional 3′UTR Editing. Cells 2021, 11, 76, 10.3390/cells11010076.
- Enrico Gaffo; Elena Boldrin; Anna Dal Molin; Silvia Bresolin; AnnaGiulia Bonizzato; Luca Trentin; Chiara Frasson; Klaus-Michael Debatin; Lueder H. Meyer; Geertruy Te Kronnie; et al.Stefania Bortoluzzi Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Scientific Reports 2019, 9, 1-12, 10.1038/s41598-019-50864-z.
- Characterization of new circular RNA products from the Pax-5 gene in B-cells
- Brandon Hannay; Pascal Dumas; Vanessa Veilleux; Nicolas LeBlanc; Gilles A. Robichaud; Discovery of Novel Non‐Coding Products of the Pax‐5 Gene and Their Clinical Significance in Lymphoid Cancers. The FASEB Journal 2018, 32, 677.23-677.23, 10.1096/fasebj.2018.32.1_supplement.677.23.
- Julia Salzman; Charles Gawad; Peter Lincoln Wang; Norman Lacayo; Patrick O. Brown; Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLOS ONE 2012, 7, e30733, 10.1371/journal.pone.0030733.
- Minghon Lu; Circular RNA: functions, applications and prospects. ExRNA 2020, 2, 1-7, 10.1186/s41544-019-0046-5.
- Laura Santer; Christian Bär; Thomas Thum; Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Molecular Therapy 2019, 27, 1350-1363, 10.1016/j.ymthe.2019.07.001.
- Amaresh Chandra Panda; Circular RNAs Act as miRNA Sponges. null 2018, 1087, 67-79, 10.1007/978-981-13-1426-1_6.
- Anne-Catherine Prats; Florian David; Leila Diallo; Emilie Roussel; Florence Tatin; Barbara Garmy-Susini; Eric Lacazette; Circular RNA, the Key for Translation. null 2020, -, -, 10.20944/preprints202010.0088.v2.
- Yun Yang; Xiaojuan Fan; Miaowei Mao; Xiaowei Song; Ping Wu; Yang Zhang; Yongfeng Jin; Yi Yang; Ling-Ling Chen; Yang Wang; et al.Catherine Cl WongXinshu XiaoZefeng Wang Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Research 2017, 27, 626-641, 10.1038/cr.2017.31.
- Sabah Nisar; Ajaz A. Bhat; Mayank Singh; Thasni Karedath; Arshi Rizwan; Sheema Hashem; Puneet Bagga; Ravinder Reddy; Farrukh Jamal; Shahab Uddin; et al.Gyan ChandDavide BedognettiWael El-RifaiMichael P. FrenneauxMuzafar A. MachaIkhlak AhmedMohammad Haris Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Frontiers in Cell and Developmental Biology 2021, 9, -, 10.3389/fcell.2021.617281.
- Giulia Fontemaggi; Chiara Turco; Gabriella Esposito; Silvia Di Agostino; New Molecular Mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers 2021, 13, 3154, 10.3390/cancers13133154.
- Characterization of new circular rna products from the pax-5 gene in b-cells
- Brandon Hannay; Pascal Dumas; Vanessa Veilleux; Nicolas LeBlanc; Gilles A. Robichaud; Discovery of Novel Non‐Coding Products of the Pax‐5 Gene and Their Clinical Significance in Lymphoid Cancers. The FASEB Journal 2018, 32, 677.23-677.23, 10.1096/fasebj.2018.32.1_supplement.677.23.
- Jennifer K. King; Nolan M. Ung; May H. Paing; Jorge R. Contreras; Michael O. Alberti; Thilini R. Fernando; Kelvin Zhang; Matteo Pellegrini; Dinesh S. Rao; Regulation of Marginal Zone B-Cell Differentiation by MicroRNA-146a. Frontiers in Immunology 2017, 7, 670, 10.3389/fimmu.2016.00670.
- Maoyi Lai; Alicia Gonzalez-Martin; Anthony B. Cooper; Hiroyo Oda; Hyun Yong Jin; Jovan Shepherd; Linling He; Jiang Zhu; David Nemazee; Maoyi Lai Alicia Gonzalez-Martin Anthony B. Cooper Hiroyo Oda Hyun Yong Jin Jovan Shepherd Linling He Jiang Zhu David Nemazee Changchun Xiao; et al. Regulation of B-cell development and tolerance by different members of the miR-17∼92 family microRNAs. Nature Communications 2016, 7, 12207, 10.1038/ncomms12207.
- James N. Psathas; Patrick J. Doonan; Pichai Raman; Bruce D. Freedman; Andy J. Minn; Andrei Thomas-Tikhonenko; The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood 2013, 122, 4220-4229, 10.1182/blood-2012-12-473090.
- Michele Dal Bo; Riccardo Bomben; Luis Hernández; Valter Gattei; The MYC/miR-17-92 axis in lymphoproliferative disorders: A common pathway with therapeutic potential. Oncotarget 2015, 6, 19381-19392, 10.18632/oncotarget.4574.
- Takahiko Yasuda; Fumihiko Hayakawa; Shingo Kurahashi; Keiki Sugimoto; Yosuke Minami; Akihiro Tomita; Tomoki Naoe; B Cell Receptor-ERK1/2 Signal Cancels PAX5-Dependent Repression of BLIMP1 through PAX5 Phosphorylation: A Mechanism of Antigen-Triggering Plasma Cell Differentiation. The Journal of Immunology 2012, 188, 6127-6134, 10.4049/jimmunol.1103039.
- Yuichiro Inagaki; Fumihiko Hayakawa; Daiki Hirano; Yuki Kojima; Takanobu Morishita; Takahiko Yasuda; Tomoki Naoe; Hitoshi Kiyoi; PAX5 tyrosine phosphorylation by SYK co-operatively functions with its serine phosphorylation to cancel the PAX5-dependent repression of BLIMP1: A mechanism for antigen-triggered plasma cell differentiation. Biochemical and Biophysical Research Communications 2016, 475, 176-181, 10.1016/j.bbrc.2016.05.067.
- Cecilia R. Kovac; Alexander Emelyanov; Mallika Singh; Nasrin Ashouian; Barbara K. Birshtein; BSAP (Pax5)-Importin α1 (Rch1) Interaction Identifies a Nuclear Localization Sequence. Journal of Biological Chemistry 2000, 275, 16752-16757, 10.1074/jbc.m001551200.
