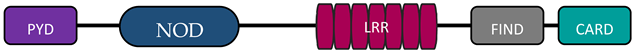Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Amina Yu and Version 1 by Magdalena Druszczyńska.
The innate immune system recognizes pathogen-associated molecular motifs through pattern recognition receptors (PRRs) that induce inflammasome assembly in macrophages and trigger signal transduction pathways, thereby leading to the transcription of inflammatory cytokine genes. Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) represent a family of cytosolic PRRs involved in the detection of intracellular pathogens such as mycobacteria or viruses.
- NLR
- antimicrobial immunity
- ssRNA virus
1. Introduction
Pattern recognition receptors (PRRs) play an important role in non-specific immunity. In a sense, these receptors provide a universal alarm signal that enables the rapid transmission of danger information and thus an immediate host defense. PRRs recognize not only pathogen components, such as lipopolysaccharide, glycolipids, and nucleic acids, but also toxic metabolic products. An important class of intracellular PRRs is the NOD-like receptor (NLR) family [1,2][1][2].
2. The NLR Family
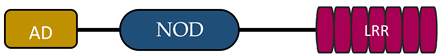
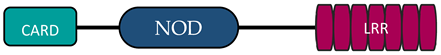
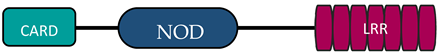
In humans, the NLR family includes at least 23 receptors that share a similar structure. Most proteins belonging to the NLR family contain three domains: an N-terminal signaling domain (pyrin domain (PYD) or caspase recruitment containing domain (CARD)), central nucleotide binding and oligomerization domain (NOD1 or NOD2), and C-terminal agonist sensing/binding domain (Leucine Rich Repeat (LRR)) [3]. Based on the N-terminal region, this family has been divided into four groups: (1) NLRP (NOD-like receptor P; P for PYD), (2) NLRC (NOD-like receptor C; C for CARD), (3) NLRA (NOD-like receptor A; A for the acidic transactivating domain), (4) NLRB (NOD-like receptor B; B for the baculovirus inhibitor of apoptosis (BIR)) (Table 1).
Table 1.
Structures of the NLR subfamilies.
Abbreviations: AD, acidic activation domain; BIR, baculovirus inhibitor of apoptosis; CARD, caspase recruitment containing domain; CIITA, class II transactivator; FIND, finder function domain; NAIP, neuronal apoptosis inhibitory protein, NLRA, NOD-like receptor A; NLRB, NOD-like receptor B; NLRC, NOD-like receptor C; NLRP, NOD-like receptor P; NOD, nucleotide-binding oligomerization domain; LRR, leucine rich repeat; PYD, pyrin domain.
NLR receptors play a key role in regulating the host innate immune response. They recognize a variety of ligands from microbial pathogens (peptidoglycan, viral RNA, flagellin), host cells (cholesterol crystals, uric acid), and environmental sources (asbestos, silica, alum).The majority of NLRs function as PRRs, detecting specific ligands and inducing inflammatory reactions, however some NLRs do not function as PRRs but respond to cytokines, such as interferons. Inflammasome platforms formed by NLRs, which function as scaffold proteins, stimulate mitogen-activated protein kinase (MAPK) and nuclear factor-κβ (NF-κβ) signaling pathways and control the activation of inflammatory caspases. In this Hereview, we discuss in, the role of three NLR molecules was discussed, NOD1, NOD2, and NLRC, in antiviral and antimicrobial immune responses, providing insight into the molecular mechanisms and their potential implications for health and disease.
2.1. NLRP
The most abundant group of NLRs is the NLRP subfamily, consisting of 14 proteins that are mostly composed of a PYD effector domain, a central NOD domain, and a C-terminal LRR domain. NLRP1 and NLRP10 have a different structure. NLRP1 protein has an additional finder function domain (FIND) and a caspase activation and recruitment domain (CARD), whereas NLRP10 lacks the LRR domain [4][8]. Recent studies have shown that autolytic proteolysis within FIND is non-specific for inflammasome activity [5][9], but the specific function of this domain is unknown. In contrast, CARD has the ability to attach caspases involved in inflammation and apoptosis (caspase 1) [6,7][10][11]. Many receptors in this subfamily (NLRP1, NLRP3, NLRP6, NLRP7, NRLP10, and NRLP12) are involved in inflammasome formation [8][12].
2.2. NLRC
The NLRC subfamily consists of six proteins, NOD1, NOD2, NLRC3, NLRC4, NLRC 5, and NLRX1—also named NOD5 or NOD9—and their common feature is the presence of the LRR and NOD domains. NLRX1 is unique because of the fact that, unlike the other NLRs, it is located in the mitochondria. The protein is referred to as NLRX1 since its N-terminus has not been completely defined [9][6]. There are different receptor structures in the NLRC subfamily. NOD1 and NLRC4 have one CARD domain and NOD2 has two CARD domains, while NLRC3, NLRC5, and NLRX1 still do not have an identified NH2 domain [10][7]. The best known proteins are NOD1 and NOD2, which recognize different bacterial and viral components [11][13]. NOD1 recognizes gamma-glutamyl diaminopimelic acid (iE-DAP), a peptidoglycan product of Gram-negative bacteria. In contrast, NOD2 recognizes muramyl dipeptide (MDP) present in the peptidoglycans of most bacteria [12,13][14][15]. Recent studies have shown that NLRs can also recognize some viruses through NOD2, which reacts with viral single-stranded RNA (ssRNA), leading to type I interferon production [14,15][16][17].
2.3. NLRA and NLRB
In the other two subfamilies (NLRA, NLRB), one member of each has now been identified, a class II transactivator (CITTA) and a neuronal apoptosis inhibitory protein (NAIP), respectively. What distinguishes these receptors from the others is the presence of an acidic activation domain (AD) in NLRA and BIR in NLRB [16,17][4][5]. CIITA activation is regulated by a number of post-translational modifications, such as acetylation, phosphorylation, and ubiquitination. CIITA activates MHC class II gene expression in different populations of antigen-presenting cells (APCs). In the absence of CIITA expression, MHC class II activation does not occur. CIITA is responsible for the expression of accessory proteins required for proper peptide presentation through MHC class II [16,18,19][4][18][19]. NAIP member, NLRB, is involved in host defence and cell survival. The characteristic feature of this protein is the inhibition of apoptosis by suppressing the activities of caspase (CASP) 3 (CASP3), CASP7, and CASP9; the autocleavage of pro-CASP9; and the cleavage of pro-CASP3 by CASP9 [16,20][4][20].
3. Structure and Expression of NOD1 and NOD2
Through sequence-homology searches, we were able to identify NOD1 and NOD2 as the first members of the NLR family will be identified. The NOD1 receptor consists of 953 amino acids and is located on chromosome 7p14-p15, while NOD2 contains 1040 amino acids and is located on human chromosome 16p21 [21,22,23][21][22][23]. While NOD1 shows a ubiquitous expression pattern, NOD2 is more strongly expressed in macrophages, dendritic cells, paneth cells, keratinocytes, epithelial intestinal cells, lung epithelial cells, oral epithelial cells, and osteoblasts ([24]). Both receptors recognize peptidoglycan residues, with NOD1 reacting with the peptidoglycan (PG) group containing gamma-glutamyl diaminopimelic acid (iE-DAP), which is mainly found in Gram-negative bacteria [13,25,26][15][25][26]. The NOD2 receptor senses MDP structures found in both Gram-positive and Gram-negative bacteria [12,27][14][27]. Recent studies suggest that NOD2 directly binds MDP with high affinity [28], and N-glycolyl MDP more strongly modulates the host response compared to N-acetyl MDP [29]. An increasing number of studies suggest that, in addition to peptidoglycan recognition, NOD1 and NOD2 may respond to danger signals or damage-associated molecular patterns (DAMPs). Endoplasmic reticulum (ER) stress contributes to inflammatory diseases such as Crohn’s disease and type 2 diabetes. ER stress induced by various microbial infections can be interpreted as a danger signal recognized by NOD1 and NOD2. Brucella abortus infection was shown to induce ER stress, which promoted NOD1/2-dependent inflammation and IL-6 production [30]. Furthermore, bacterial effector proteins can activate NOD1 and NOD2. Shigella flexneri effector proteins, such as OspB and IpgB2, and Salmonella enterica serotype, Typhimurium SipA and SopE, induce NF-κB activation in a peptidoglycan-independent, but NOD1- or NOD2-dependent, manner [31,32,33][31][32][33]. Recent studies have shown that NLRs can also recognize certain viruses through NOD2, which reacts with viral ssRNA, leading to type I interferon production [14,15][16][17]. Finally, NOD1 and NOD2 signaling leads to the activation of host defense mechanisms, such as the production of pro-inflammatory cytokines and antimicrobial molecules [34,35][34][35]. NOD1 and NOD2 are cytoplasmic receptors, but their activation depends on their location in endosomes and in the plasma membrane [36,37,38][36][37][38]. The recruitment of NOD1 and NOD2 to the cell membrane occurs through interaction with the actin cytoskeleton [39,40][39][40]. Rho GTPases, which are responsible for regulating the actin cytoskeleton, are known to be able to interact with and activate NOD1 [32]. Furthermore, NOD2 binds to regulators of the actin cytoskeleton, GTPase Arf [41]. Moreover, transport to the cell membrane requires lipid modification (S-palmitoylation) for recruitment to the cell membrane [42].
Upon recognition of the appropriate agonist by NOD1 and NOD2 receptors, activation of pathways whose primary targets are MAPK, caspase-1, and NF-κΒ occurs. These activations lead to the transcription of relevant genes and the production of inflammatory mediators. The activation of NOD1 and NOD2 signaling pathways is regulated by post-translational modifications, such as phosphorylation and ubiquitination. The recognition of iE-DAP and MADP by NOD1 and NOD2, respectively, leads to a change in receptor conformation and exposure of the CARD domain, which recruits and activates receptor-interacting serine/threonine kinase 2 (RIP2) [21,43,44][21][43][44]. The activation of RIP2 kinase is necessary to activate the inflammatory signaling pathway associated with NOD1 and NOD2 receptors [45]. In the next step, RIP2 can lead to the ubiquitination of the essential modulator of NF-κB (NEMO) and activation of the IKK (IκB kinase) complex. The IKK complex phosphorylates the inhibitor of kappaB (IκBα), which is then degraded, and the released NF-κB is translocated to the cell nucleus. In the cell nucleus, NF-κB dimers bind to kappaB (κB) elements, activating pro-inflammatory cytokines and factors responsible for immune cells, among others [46,47,48,49][46][47][48][49] (Figure 1).
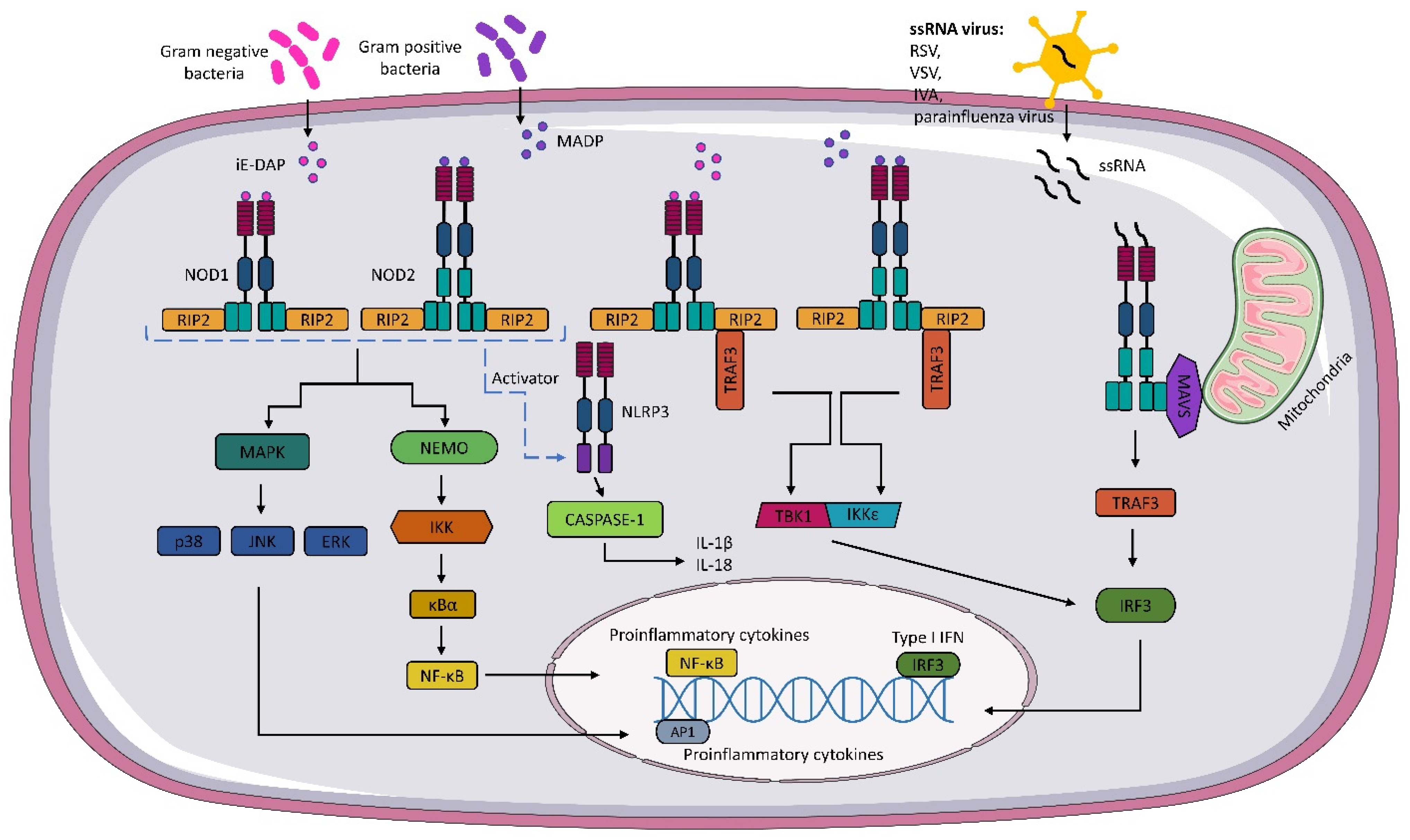
Figure 1. NOD1 and NOD2 signaling pathway. Gamma-glutamyl diaminopimelic acid (iE-DAP) and muramyl dipeptide (MDP) activate the nucleotide-binding oligomerization domains 1/2 (NOD1) and (NOD2), respectively. Activation of NODs leads to the recruitment of receptor-interacting serine/threonine kinase (RIP2). In the next step, activated RIP2 can lead to ubiquitination of the essential modulator of NF-κB (NEMO) and activation of the IKK (IκB kinase) complex. The activated IKK complex phosphorylates the inhibitor of kappaB (IκBα). The activated IKK complex phosphorylates the kappaB inhibitor (IκBα), leading to the release of NF-κB, which, after translocation to the cell nucleus, binds to kappaB (κB) elements, thereby activating pro-inflammatory cytokines. NOD1 and NOD2 also activate mitogen-activated protein kinases (MAPKs), such as p38, c-Jun N-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK). NOD1 and NOD2 also interact with the NLRP3 inflammasome, which leads to caspase-1 activation and IL-18 and IL-1β production. Activation of NOD1 and NOD2 results in the formation of a TBK1 and the inhibitor of the nuclear factor kappaB kinase (IKKε) complex, which leads to the expression of type I IFNs. As well, through the interaction of NOD2 with mitochondrial antiviral signaling protein MAVS, the expression of IFN I genes occurs.
The formation of nodosome complexes can also lead to the activation of one of the three major MAP kinase families: p38, c-Jun N-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK). As a result, the MAPK pathway is activated, and p38, JNK, and ERK translocate to the nucleus and phosphorylate AP-1 transcription factors responsible for the expression of pro-inflammatory cytokines and chemokines [34,50,51][34][50][51] (Figure 1).
In addition to the pathways mentioned above, NOD 1 and NOD2 receptors can also activate the interferon signaling pathway. Sabbah et al. showed that NOD2 not only recognizes MDP, but also interacts with viral ssRNA [14][16]. The researchers revealed that RSV activates NOD2 through the mitochondrial antiviral signaling protein (MAVS), which leads to the activation of the regulatory transcription factor, interferon 3 (IRF3). In subsequent steps, IRF3 migrates to the nucleus and activates type I IFN (IFNα/β) genes. Interestingly, the IFN pathway is also activated during bacterial infections. The mechanism is not fully understood, but it has been shown that upon binding of bacterial ligands, conformational changes of NOD1 and NOD2 occur, followed by the formation of the RIP2-TRAF3 complex, which recruits TANK shunt kinase 1 (TBK1) and the inhibitor of nuclear factor kappaB kinase (IKKε). These two kinases form a TBK1-IKKε complex that drives IRF3-dependent expression of IFNβ and type I IFN genes [52,53][52][53] (Figure 1).
It is still not entirely clear how the NOD1 and NOD2 signaling pathways are regulated. Several studies have shown that certain molecules can positively or negatively affect NOD1 and NOD2 activation pathways. GRIM-19 protein and vimentin have been described as positive regulators of this process. They are required for NF-κB activation after MDP recognition by NOD2 [54,55][54][55]. A negative regulator of NOD signaling is the Erbin protein, which, when bound to NOD2, inhibits MDP-induced signaling [56]. Moreover, overexpression of Centaurin β-1 (CENTβ1), a GTPase-activating protein, inhibits NOD1- and NOD2-dependent expression of NF-κB [57].
4. Structure and Expression NLRC5
Among NLR members, NLRC5 contains the largest number of LRRs, consisting of 713 amino acids, making it the largest protein in this receptor family [58]. The complete human NLRC5 gene consists of 1866 amino acids and is located at locus 16q13 [59]. NLRC5 responds to viral components and lipopolysaccharides, and is highly sensitive to interferon γ (IFN-γ) [58,60,61][58][60][61]. NLRC5 has a structure similar to the other receptors belonging to the NLR family; however, the N-terminal CARD of NLRC5 is characterized by the folding of the death domain, making it an atypical CARD domain [62]. Furthermore, the NOD domain of the NLRC5 receptor contains the Walker A and Walker B regions, which are required for the binding and hydrolysis of nucleotide triphosphate, respectively. The Walker A motif is essential for NLRC5 migration into the nucleus and, consequently, for the transactivation activity of major histocompatibility complex (MHC) class I genes [63,64][63][64]. The structure of NLRC5 is highly conserved in many mammalian species; the homology of human NLRC5 to mice NLRC5 is 64% [65], suggesting that NLRC5 shows similar functions in different organisms.NLRC5 is highly expressed in immune cells and tissues, such as the spleen, lymph nodes, bone marrow, thymus, intestine, lung, and bone marrow [59,60,65][59][60][65]. NLRC5 has different functions depending on its localization. In the cytoplasm, NLRC5 can inhibit nuclear factor kappa B (NF-κB) signaling by interacting with IκB kinase alpha/beta (IKKα/β) so that NEMO does not bind to IKK and autophosphorylation and kinase activity are inhibited [59,65][59][65]. NLRC5 in the cytoplasm is involved in the regulation of type I IFN, but data on the effect of this receptor on IFN I are conflicting. Some studies suggest that NLRC5 interacts with RIG-I and MAVS, resulting in an enhanced IFN I response. (Figure 2) However, there are also reports suggesting that NLRC5 inhibits the IFN I response [65,66,67][65][66][67]. The formation of the inflammasome is important in the recognition of the infectious agent and in modulating the host immune response. It has been speculated that NLRC5 may lead to the activation of the inflammasome and even to the formation of its own inflammasome [68]. NLRC5 is involved in the activation of the NLRP3 inflammasome. The overexpression of NLRC5 leads to the increased activation of caspase-1, which converts pro-IL-1β into active IL-1β [67,68][67][68] (Figure 2). Tong et al. and Kumar et al. observed similar levels of IL-1β secretion in wild-type and NLRC5-/- mice [67,69][67][69]. Recent studies suggest that NLRC5 interacts with tumors through multiple pathways, such as β-catenin, TGF-β, and Akt [70,71,72][70][71][72]. It is worth noting that most of the presented functions of NLRC5 are still unclear and debatable, so further studies on this receptor are needed.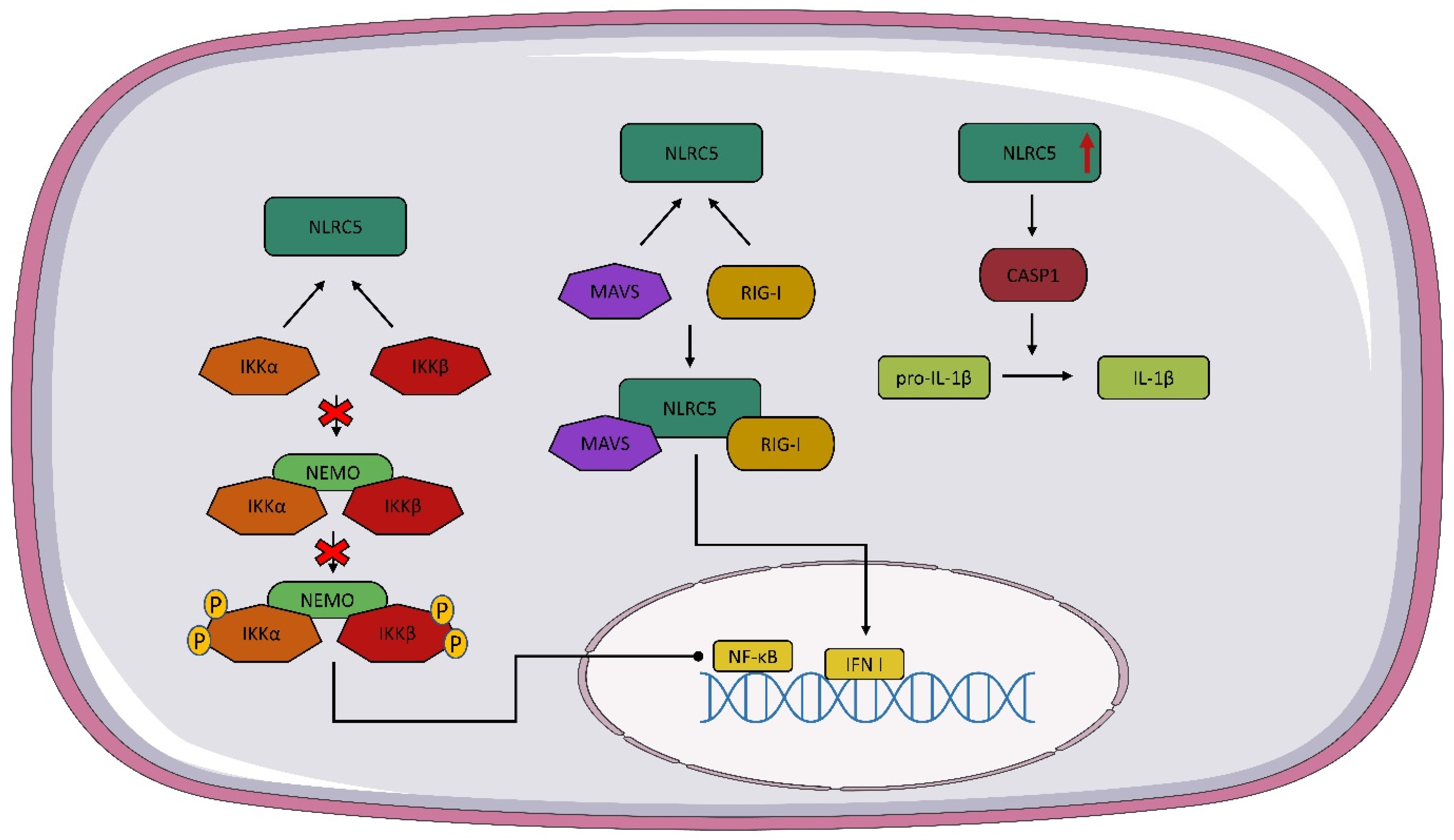
Figure 2. NLRC5 signaling pathway. As a result of the interaction of NOD-like receptor C5 NLRC5 with IκB kinase alpha/beta (IKKα/β), the NEMO IKKα/β complex is not formed and nuclear factor kappa B (NF-κB) expression is stopped. The interaction of NLRC5 with mitochondrial antiviral signaling protein (MAVS) and RIG-I leads to the formation of a complex of NLRC5 plus MAVS and RIG-I, which contributes to the expression of IFN I. Overexpression of NLCR5 activates caspase 1 (CASP1), which converts pro-IL-1β to active IL-1β.
References
- Fritz, J.H.; Ferrero, R.L.; Philpott, D.J.; Girardin, S.E. Nod-like Proteins in Immunity, Inflammation and Disease. Nat. Immunol. 2006, 7, 1250–1257.
- Zhong, Y.; Kinio, A.; Saleh, M. Functions of NOD-Like Receptors in Human Diseases. Front. Immunol. 2013, 4, 333.
- Zeromski, J.; Kierepa, A.; Boruczkowski, M.; Kowala-Piaskowska, A.; Mozer-Lisewska, I. The Role Of Nod-Like Receptors In Immunobiology And Medicine. Adv. Cell Biol. 2017, 44, 201–212.
- Chen, L.; Cao, S.Q.; Lin, Z.M.; He, S.J.; Zuo, J.P. NOD-like Receptors in Autoimmune Diseases. Acta Pharmacol. Sin. 2021, 42, 1742–1756.
- Bauernfeind, F.; Ablasser, A.; Bartok, E.; Kim, S.; Schmid-Burgk, J.; Cavlar, T.; Hornung, V. Inflammasomes: Current Understanding and Open Questions. Cell. Mol. Life Sci. 2010, 68, 765–783.
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. Nod-like Receptors: Versatile Cytosolic Sentinels. Physiol. Rev. 2015, 95, 149–178.
- Proell, M.; Riedl, S.J.; Fritz, J.H.; Rojas, A.M.; Schwarzenbacher, R. The Nod-Like Receptor (NLR) Family: A Tale of Similarities and Differences. PLoS ONE 2008, 3, e2119.
- Macdonald, J.A.; Wijekoon, C.P.; Liao, K.C.; Muruve, D.A. Biochemical and Structural Aspects of the ATP-Binding Domain in Inflammasome-Forming Human NLRP Proteins. IUBMB Life 2013, 65, 851–862.
- Finger, J.N.; Lich, J.D.; Dare, L.C.; Cook, M.N.; Brown, K.K.; Duraiswamis, C.; Bertin, J.J.; Gough, P.J. Autolytic Proteolysis within the Function to Find Domain (FIIND) Is Required for NLRP1 Inflammasome Activity. J. Biol. Chem. 2012, 287, 25030–25037.
- Boucher, D.; Monteleone, M.; Coll, R.C.; Chen, K.W.; Ross, C.M.; Teo, J.L.; Gomez, G.A.; Holley, C.L.; Bierschenk, D.; Stacey, K.J.; et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 2018, 3, 827–840.
- Bronner, D.N.; Abuaita, B.H.; Chen, X.; Fitzgerald, K.A.; Nuñez, G.; He, Y.; Yin, X.M.; O’Riordan, M.X. Endoplasmic Reticulum Stress Activates the Inflammasome via NLRP3- and Caspase-2-Driven Mitochondrial Damage. Immunity 2015, 43, 451–462.
- Platnich, J.M.; Muruve, D.A. NOD-like Receptors and Inflammasomes: A Review of Their Canonical and Non-Canonical Signaling Pathways. Arch. Biochem. Biophys. 2019, 670, 4–14.
- Bertsche, U.; Mayer, C.; Götz, F.; Gust, A.A. Peptidoglycan Perception—Sensing Bacteria by Their Common Envelope Structure. Int. J. Med. Microbiol. 2015, 305, 217–223.
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 Is a General Sensor of Peptidoglycan through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872.
- Chamaillard, M.; Hashimoto, M.; Horie, Y.; Masumoto, J.; Qiu, S.; Saab, L.; Ogura, Y.; Kawasaki, A.; Fukase, K.; Kusumoto, S.; et al. An Essential Role for NOD1 in Host Recognition of Bacterial Peptidoglycan Containing Diaminopimelic Acid. Nat. Immunol. 2003, 4, 702–707.
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of Innate Immune Antiviral Responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080.
- Kim, J.; Yang, Y.L.; Jang, Y.S. Human β-Defensin 2 Is Involved in CCR2-Mediated Nod2 Signal Transduction, Leading to Activation of the Innate Immune Response in Macrophages. Immunobiology 2019, 224, 502–510.
- Leibund Gut-Landmann, S.; Waldburger, J.M.; Krawczyk, M.; Otten, L.A.; Suter, T.; Fontana, A.; Acha-Orbea, H.; Reith, W. Mini-Review: Specificity and Expression of CIITA, the Master Regulator of MHC Class II Genes. Eur. J. Immunol. 2004, 34, 1513–1525.
- Accolla, R.S.; Ramia, E.; Tedeschi, A.; Forlani, G. CIITA-Driven MHC Class II Expressing Tumor Cells as Antigen Presenting Cell Performers: Toward the Construction of an Optimal Anti-Tumor Vaccine. Front. Immunol. 2019, 10, 1806.
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like Receptors: Major Players (and Targets) in the Interface between Innate Immunity and Cancer. Biosci. Rep. 2019, 39, 1–21.
- Inohara, N.; Koseki, T.; del Peso, L.; Hu, Y.; Yee, C.; Chen, S.; Carrio, R.; Merino, J.; Liu, D.; Ni, J.; et al. Nod1, an Apaf-1-like Activator of Caspase-9 and Nuclear Factor-KappaB. J. Biol. Chem. 1999, 274, 14560–14567.
- Ogura, Y.; Inohara, N.; Benito, A.; Chen, F.F.; Yamaoka, S.; Nunez, G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 2001, 276, 4812–4818.
- Sidiq, T.; Yoshihama, S.; Downs, I.; Kobayashi, K.S. Nod2: A Critical Regulator of Ileal Microbiota and Crohn’s Disease. Front. Immunol. 2016, 7, 367.
- Moreira, L.O.; Zamboni, D.S. NOD1 and NOD2 Signaling in Infection and Inflammation. Front. Immunol. 2012, 3, 328.
- Hasegawa, M.; Yang, K.; Hashimoto, M.; Park, J.H.; Kim, Y.G.; Fujimoto, Y.; Nuñez, G.; Fukase, K.; Inohara, N. Differential Release and Distribution of Nod1 and Nod2 Immunostimulatory Molecules among Bacterial Species and Environments. J. Biol. Chem. 2006, 281, 29054–29063.
- Girardin, S.E.; Boneca, I.G.; Carneiro, L.A.M.; Antignac, A.; Jéhanno, M.; Viala, J.; Tedin, K.; Taha, M.K.; Labigne, A.; Zähringer, U.; et al. Nod1 Detects a Unique Muropeptide from Gram-Negative Bacterial Peptidoglycan. Science 2003, 300, 1584–1587.
- Inohara, N.; Ogura, Y.; Fontalba, A.; Gutierrez, O.; Pons, F.; Crespo, J.; Fukase, K.; Inamura, S.; Kusumoto, S.; Hashimoto, M.; et al. Host Recognition of Bacterial Muramyl Dipeptide Mediated through NOD2: Implications for Crohn′s disease. J. Biol. Chem. 2003, 278, 5509–5512.
- Grimes, C.L.; Ariyananda, L.D.Z.; Melnyk, J.E.; O’Shea, E.K. The Innate Immune Protein Nod2 Binds Directly to MDP, a Bacterial Cell Wall Fragment. J. Am. Chem. Soc. 2012, 134, 13535–13537.
- Coulombe, F.; Divangahi, M.; Veyrier, F.; de Léséleuc, L.; Gleason, J.L.; Yang, Y.; Kelliher, M.A.; Pandey, A.K.; Sassetti, C.M.; Reed, M.B.; et al. Increased NOD2-Mediated Recognition of N-Glycolyl Muramyl Dipeptide. J. Exp. Med. 2009, 206, 1709.
- Keestra-Gounder, A.M.; Byndloss, M.X.; Seyffert, N.; Young, B.M.; Chávez-Arroyo, A.; Tsai, A.Y.; Cevallos, S.A.; Winter, M.G.; Pham, O.H.; Tiffany, C.R.; et al. NOD1/NOD2 Signaling Links ER Stress with Inflammation. Nature 2016, 532, 394.
- Marijke Keestra, A.; Winter, M.G.; Klein-Douwel, D.; Xavier, M.N.; Winter, S.E.; Kim, A.; Tsolis, R.M.; Bäumler, A.J. A Salmonella Virulence Factor Activates the NOD1/NOD2 Signaling Pathway. mBio 2011, 2, e00266-11.
- Keestra, A.M.; Winter, M.G.; Auburger, J.J.; Fräßle, S.P.; Xavier, M.N.; Winter, S.E.; Kim, A.; Poon, V.; Ravesloot, M.M.; Waldenmaier, J.F.T.; et al. Manipulation of Small Rho GTPases Is a Pathogen-Induced Process Detected by Nod1. Nature 2013, 496, 233.
- Fukazawa, A.; Alonso, C.; Kurachi, K.; Gupta, S.; Lesser, C.F.; McCormick, B.A.; Reinecker, H.C. GEF-H1 Mediated Control of NOD1 Dependent NF-ΚB Activation by Shigella Effectors. PLoS Pathog. 2008, 4, 1000228.
- Kobayashi, K.S.; Chamaillard, M.; Ogura, Y.; Henegariu, O.; Inohara, N.; Nuñez, G.; Flavell, R.A. Nod2-Dependent Regulation of Innate and Adaptive Immunity in the Intestinal Tract. Science 2005, 307, 731–734.
- Masumoto, J.; Yang, K.; Varambally, S.; Hasegawa, M.; Tomlins, S.A.; Qiu, S.; Fujimoto, Y.; Kawasaki, A.; Foster, S.J.; Horie, Y.; et al. Nod1 Acts as an Intracellular Receptor to Stimulate Chemokine Production and Neutrophil Recruitment in Vivo. J. Exp. Med. 2006, 203, 203–213.
- Nakamura, N.; Lill, J.R.; Phung, Q.; Jiang, Z.; Bakalarski, C.; de Mazière, A.; Klumperman, J.; Schlatter, M.; Delamarre, L.; Mellman, I. Endosomes Are Specialized Platforms for Bacterial Sensing and NOD2 Signalling. Nature 2014, 509, 240–244.
- Irving, A.T.; Mimuro, H.; Kufer, T.A.; Lo, C.; Wheeler, R.; Turner, L.J.; Thomas, B.J.; Malosse, C.; Gantier, M.P.; Casillas, L.N.; et al. The Immune Receptor NOD1 and Kinase RIP2 Interact with Bacterial Peptidoglycan on Early Endosomes to Promote Autophagy and Inflammatory Signaling. Cell Host Microbe 2014, 15, 623–635.
- Travassos, L.H.; Carneiro, L.A.M.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhes, J.G.; Yuan, L.; Soares, F.; Chea, E.; le Bourhis, L.; et al. Nod1 and Nod2 Direct Autophagy by Recruiting ATG16L1 to the Plasma Membrane at the Site of Bacterial Entry. Nat. Immunol. 2010, 11, 55–62.
- Kufer, T.A.; Kremmer, E.; Adam, A.C.; Philpott, D.J.; Sansonetti, P.J. The Pattern-Recognition Molecule Nod1 Is Localized at the Plasma Membrane at Sites of Bacterial Interaction. Cell Microbiol. 2008, 10, 477–486.
- Legrand-Poels, S.; Kustermans, G.; Bex, F.; Kremmer, E.; Kufer, T.A.; Piette, J. Modulation of Nod2-Dependent NF-KappaB Signaling by the Actin Cytoskeleton. J. Cell Sci. 2007, 120, 1299–1310.
- Wang, Y.C.; Westcott, N.P.; Griffin, M.E.; Hang, H.C. Peptidoglycan Metabolite Photoaffinity Reporters Reveal Direct Binding to Intracellular Pattern Recognition Receptors and Arf GTPases. ACS Chem. Biol. 2019, 14, 405.
- Lu, Y.; Zheng, Y.; Coyaud, É.; Zhang, C.; Selvabaskaran, A.; Yu, Y.; Xu, Z.; Weng, X.; Chen, J.S.; Meng, Y.; et al. Palmitoylation of NOD1 and NOD2 Is Required for Bacterial Sensing. Science 2019, 366, 460–467.
- Inohara, N.; Chamaillard, M.; McDonald, C.; Nuñez, G. NOD-LRR proteins: Role in Host-Microbial Interactions and Inflammatory Disease. Annu. Rev. Biochem. 2005, 74, 355–383.
- Nembrini, C.; Kisielow, J.; Shamshiev, A.T.; Tortola, L.; Coyle, A.J.; Kopf, M.; Marsland, B.J. The Kinase Activity of Rip2 Determines Its Stability and Consequently Nod1- and Nod2-Mediated Immune Responses. J. Biol. Chem. 2009, 284, 19183–19188.
- Hrdinka, M.; Schlicher, L.; Dai, B.; Pinkas, D.M.; Bufton, J.C.; Picaud, S.; Ward, J.A.; Rogers, C.; Suebsuwong, C.; Nikhar, S.; et al. Small Molecule Inhibitors Reveal an Indispensable Scaffolding Role of RIPK2 in NOD2 Signaling. EMBO J. 2018, 37, e99372.
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific Recognition of Linear Ubiquitin Chains by NEMO Is Important for NF-KappaB Activation. Cell 2009, 136, 1098–1109.
- Jiang, X.; Chen, Z.J. The Role of Ubiquitylation in Immune Defence and Pathogen Evasion. Nat. Rev. Immunol. 2011, 12, 35–48.
- Abbott, D.W.; Wilkins, A.; Asara, J.M.; Cantley, L.C. The Crohn’s Disease Protein, NOD2, Requires RIP2 in Order to Induce Ubiquitinylation of a Novel Site on NEMO. Curr. Biol. 2004, 14, 2217–2227.
- Pellegrini, E.; Desfosses, A.; Wallmann, A.; Schulze, W.M.; Rehbein, K.; Mas, P.; Signor, L.; Gaudon, S.; Zenkeviciute, G.; Hons, M.; et al. RIP2 Filament Formation Is Required for NOD2 Dependent NF-ΚB Signalling. Nat. Commun. 2018, 9, 1–19.
- Girardin, S.E.; Tournebize, R.; Mavris, M.; Page, A.L.; Li, X.; Stark, G.R.; Bertin, J.; Distefano, P.S.; Yaniv, M.; Sansonetti, P.J.; et al. CARD4/Nod1 Mediates NF-KappaB and JNK Activation by Invasive Shigella Flexneri. EMBO Rep. 2001, 2, 736–742.
- Park, J.-H.; Kim, Y.-G.; Shaw, M.; Kanneganti, T.-D.; Fujimoto, Y.; Fukase, K.; Inohara, N.; Núñez, G. Nod1/RICK and TLR Signaling Regulate Chemokine and Antimicrobial Innate Immune Responses in Mesothelial Cells. J. Immunol. 2007, 179, 514–521.
- Stockinger, S.; Reutterer, B.; Schaljo, B.; Schellack, C.; Brunner, S.; Materna, T.; Yamamoto, M.; Akira, S.; Taniguchi, T.; Murray, P.J.; et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 2004, 173, 7416–7425.
- Allison, C.C.; Ferrand, J.; McLeod, L.; Hassan, M.; Kaparakis-Liaskos, M.; Grubman, A.; Bhathal, P.S.; Dev, A.; Sievert, W.; Jenkins, B.J.; et al. Nucleotide Oligomerization Domain 1 Enhances IFN-γ Signaling in Gastric Epithelial Cells during Helicobacter pylori Infection and Exacerbates Disease Severity. J. Immunol. 2013, 190, 3706–3715.
- Barnich, N.; Hisamatsu, T.; Aguirre, J.E.; Xavier, R.; Reinecker, H.C.; Podolsky, D.K. GRIM-19 Interacts with Nucleotide Oligomerization Domain 2 and Serves as Downstream Effector of Anti-Bacterial Function in Intestinal Epithelial Cells. J. Biol. Chem. 2005, 280, 19021–19026.
- Stevens, C.; Henderson, P.; Nimmo, E.R.; Soares, D.C.; Dogan, B.; Simpson, K.W.; Barrett, J.C.; Wilson, D.C.; Satsangi, J. The Intermediate Filament Protein, Vimentin, Is a Regulator of NOD2 Activity. Gut 2013, 62, 695–707.
- McDonald, C.; Chen, F.F.; Ollendorff, V.; Ogura, Y.; Marchetto, S.; Lécine, P.; Borg, J.P.; Nuñez, G. A Role for Erbin in the Regulation of Nod2-Dependent NF-ΚB Signaling. J. Biol. Chem. 2005, 280, 40301–40309.
- Yamamoto-Furusho, J.K.; Barnich, N.; Xavier, R.; Hisamatsu, T.; Podolsky, D.K. Centaurin Β1 Down-Regulates Nucleotide-Binding Oligomerization Domains 1- and 2-Dependent NF-ΚB Activation. J. Biol. Chem. 2006, 281, 36060–36070.
- Kuenzel, S.; Till, A.; Winkler, M.; Häsler, R.; Lipinski, S.; Jung, S.; Grötzinger, J.; Fickenscher, H.; Schreiber, S.; Rosenstiel, P. The Nucleotide-Binding Oligomerization Domain-Like Receptor NLRC5 Is Involved in IFN-Dependent Antiviral Immune Responses. J. Immunol. 2010, 184, 1990–2000.
- Benko, S.; Magalhaes, J.G.; Philpott, D.J.; Girardin, S.E. NLRC5 Limits the Activation of Inflammatory Pathways. J. Immunol. 2010, 185, 1681–1691.
- Neerincx, A.; Lautz, K.; Menning, M.; Kremmer, E.; Zigrino, P.; Hösel, M.; Büning, H.; Schwarzenbacher, R.; Kufer, T.A. A Role for the Human Nucleotide-Binding Domain, Leucine-Rich Repeat-Containing Family Member NLRC5 in Antiviral Responses. J. Biol. Chem. 2010, 285, 26223–26232.
- Wang, Y.; Huang, C.; Bian, E.; Lei, T.; Lv, X.; Li, J. NLRC5 Negatively Regulates Inflammatory Responses in LPS-Induced Acute Lung Injury through NF-ΚB and P38 MAPK Signal Pathways. Toxicol. Appl. Pharmacol. 2020, 403, 115150.
- Zhang, L.; Jiao, C.; Liu, L.; Wang, A.; Tang, L.; Ren, Y.; Huang, P.; Xu, J.; Mao, D.; Liu, L. NLRC5: A Potential Target for Central Nervous System Disorders. Front. Immunol. 2021, 12, 2451.
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.H.; Liu, Y.J.; Bayir, E.; Iliopoulos, D.; van den Elsen, P.J.; Kobayashi, K.S. NLR Family Member NLRC5 Is a Transcriptional Regulator of MHC Class I Genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799.
- Neerincx, A.; Rodriguez, G.M.; Steimle, V.; Kufer, T.A. NLRC5 Controls Basal MHC Class I Gene Expression in an MHC Enhanceosome-Dependent Manner. J. Immunol. 2012, 188, 4940–4950.
- Cui, J.; Zhu, L.; Xia, X.; Wang, H.Y.; Legras, X.; Hong, J.; Ji, J.; Shen, P.; Zheng, S.; Chen, Z.J.; et al. NLRC5 Negatively Regulates the NF-ΚB and Type I Interferon Signaling Pathways and Antiviral Immunity. Cell 2010, 141, 483.
- Ranjan, P.; Singh, N.; Kumar, A.; Neerincx, A.; Kremmer, E.; Cao, W.; Davis, W.G.; Katz, J.M.; Gangappa, S.; Lin, R.; et al. NLRC5 Interacts with RIG-I to Induce a Robust Antiviral Response against Influenza Virus Infection. Eur. J. Immunol. 2015, 45, 758–772.
- Kumar, H.; Pandey, S.; Zou, J.; Kumagai, Y.; Takahashi, K.; Akira, S.; Kawai, T. NLRC5 Deficiency Does Not Influence Cytokine Induction by Virus and Bacteria Infections. J. Immunol. 2011, 186, 994–1000.
- Davis, B.K.; Roberts, R.A.; Huang, M.T.; Willingham, S.B.; Conti, B.J.; Brickey, W.J.; Barker, B.R.; Kwan, M.; Taxman, D.J.; Accavitti-Loper, M.-A.; et al. Cutting Edge: NLRC5-Dependent Activation of the Inflammasome. J. Immunol. 2011, 186, 1333–1337.
- Tong, Y.; Cui, J.; Li, Q.; Zou, J.; Wang, H.Y.; Wang, R.F. Enhanced TLR-Induced NF-ΚB Signaling and Type I Interferon Responses in NLRC5 Deficient Mice. Cell Res. 2012, 22, 822–835.
- He, Y.; Li, M.; Zhang, X.; Meng, X.; Huang, C.; Li, J. NLRC5 Promotes Cell Proliferation via Regulating the AKT/VEGF-A Signaling Pathway in Hepatocellular Carcinoma. Toxicology 2016, 359–360, 47–57.
- Peng, Y.; He, Y.; Chen, C.; Xu, T.; Li, L.; Ni, M.; Meng, X.; Huang, C.; Li, J. NLRC5 Regulates Cell Proliferation, Migration and Invasion in Hepatocellular Carcinoma by Targeting the Wnt/β-Catenin Signaling Pathway. Cancer Lett. 2016, 376, 10–21.
- Ma, H.L.; Zhao, X.F.; Chen, G.Z.; Fang, R.H.; Zhang, F.R. Silencing NLRC5 Inhibits Extracellular Matrix Expression in Keloid Fibroblasts via Inhibition of Transforming Growth Factor-Β1/Smad Signaling Pathway. Biomed. Pharmacother. 2016, 83, 1016–1021.
More