Genomics is the cornerstone of breeding, and studies based on whole-genome sequencing and genome-wide association study have greatly driven forward genomics-assisted breeding in many crops. Lotus (Nelumbo nucifera), under the Nelumbonaceae family, is one of the relict plants possessing important scientific research and economic values. As a basal eudicot species, lotus plays an essential role in studying plant evolution and phylogeny. It is adapted to the aquatic environment, while its relatives are shrubs or trees living on land. Water lily lies at the phylogenetic position of the base angiosperm and has similar living conditions and flowers. However, its genomes are vastly different. Lotus has unique features such as water-repellent self-cleaning function, multi-seed production, and flower thermogenesis, which may relate to flower protogyny or provide a warm environment for pollination.
- lotus
- genome
- variant
- germplasm
- breeding
- omics
1. Introduction
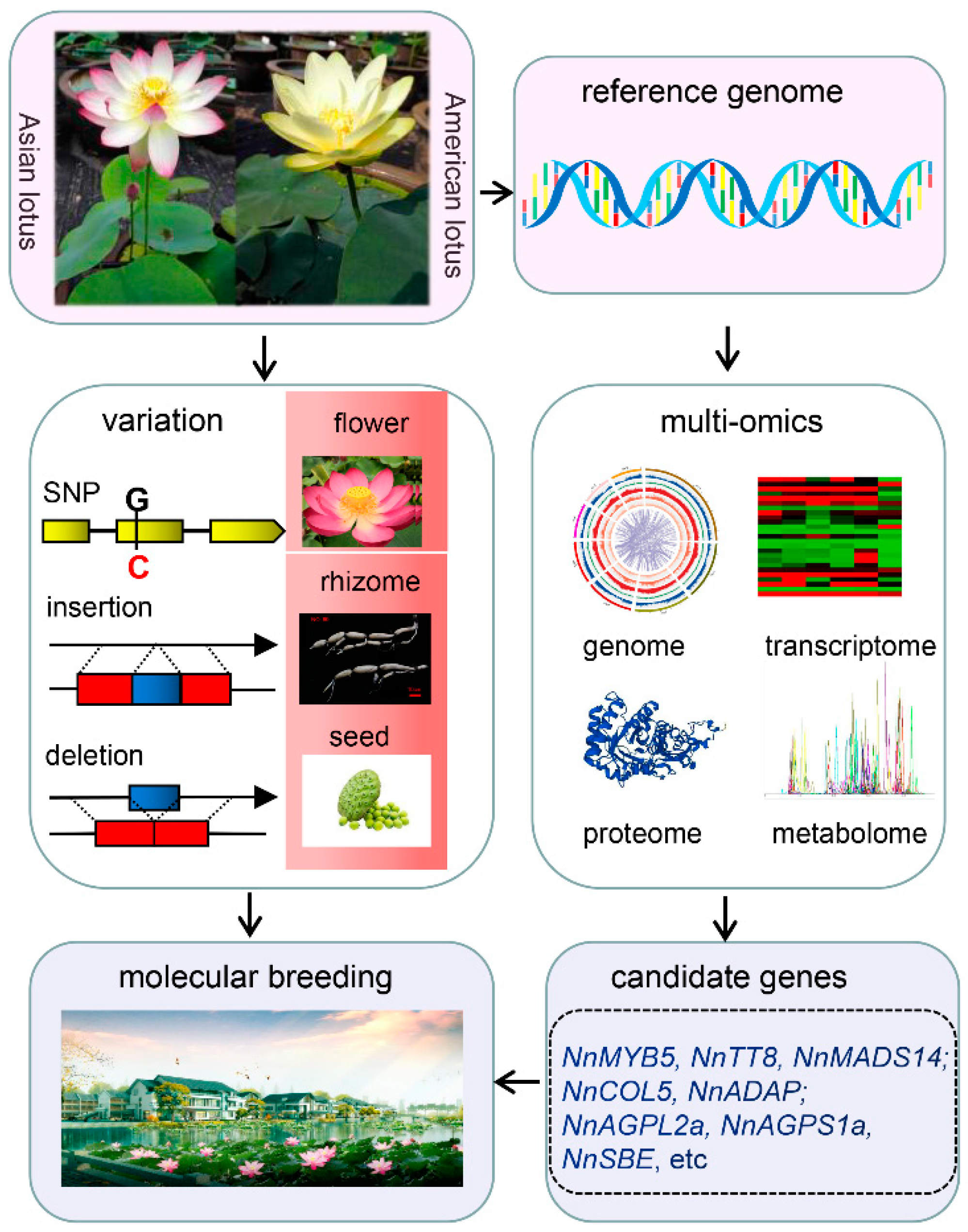
Lotus is one of the relict plants retaining the original morphology of its ancestors, as well as Ginkgo biloba, Liriodendron, and Metasequoia glyptostroboides. It belongs to the Nelumbo genus of the Nelumbonaceae family, which includes two species, namely Asian lotus (Nelumbo nucifera Gaertn.) and American lotus (Nelumbo lutea Pear.). The two species are named for their different geographical distributions. Asian lotus is mainly distributed in Asia and the north of Oceania, while American lotus is distributed in North America and South America. The plant morphology differs between them. Asian lotus is a tall plant, with oval leaves and seeds, and red or white flower colors, whereas American lotus is a short plant, nearly round and with dark green leaves, spherical seeds, and yellow flowers [16][1]. There is no strict reproductive isolation between them, and the life cycles are similar at about five months. Asian lotus is commonly called lotus and has more than 3000 years of cultivation history as a horticultural crop [17][2]. Lotus seeds and rhizomes have rich nutritional value and unique health-care function. Lotus seeds contain starch, proteins, amino acids, polysaccharides, polyphenols, alkaloids, and mineral elements. Lotus rhizome has a high vitamin C content. During the long period of domestication and artificial selection, about 4500 lotus cultivars have been obtained up till the present [18][3]. These cultivars have been planted to produce edible vegetables, snacks, beverages, restorative materials, and ornamental flowers, which impact human life and economic development. The lotus industry is also important for rural revitalization in the Yangtze River, Pearl River, and Huang Huai river basins. The cultivated lotus is generally divided into rhizome lotus, seed lotus, and flower lotus based on their different usage. The notable feature of the rhizome lotus is the enlarged rhizome but with few flowers. It can be divided into power and crisp type according to the taste of the rhizome. Different varieties were bred to meet the taste of the different regions of people or for further usage. The main breeding goal of rhizome lotus is to improve the yield and quality of the rhizome. Seed lotus is mainly for lotus seed production, with high yield, good quality, and disease resistance being the breeding goals. Flower lotus is preferred for ornamental use, and it has distinct flower colors and shapes. During long cultivation, ornamental lotus with different flower morphologies were obtained, including few-petaled, double-petaled, petaloidy, and thousand-petalled flowers. Red, pink, yellow, and white are the main flower colors. Currently, the breeding objective is mainly aimed at flower shape and color, yield or quality of lotus seed and rhizome, and wide adaptability.
2. Sequencing, Assembly and Annotation of Lotus Genome
| Items | Year | Sequencing Technology | Final Assembly (Mb) | Contig N50 | Number of Genes | Repeat Sequences | Ref. | |
|---|---|---|---|---|---|---|---|---|
| China Antique v1.0 | 2013 | Illumina, 454 | 804 | 38.8 Kb | 26685 | 57% | [20] | [4] |
| Taizi | 2013 | Illumina Hiseq2000 | 792 | 39.3 Kb | 40348 | 49.48% (TEs) | [21] | [5] |
| China Antique v2.0 | 2020 | Pacbio Sequel, Illumina | 821.2 | 484.3 Kb | 32124 | 58.50% | [23] | [7] |
| Taikonglian NO.3 | 2022 | Nanopore | 807 | 5.1 Mb | 28274 | 63.11% | [24] | [8] |
| American lotus | 2022 | Pacbio RSII, Hi-C | 843 | 1.34 Mb | 31382 | 81.00% | [25] | [9] |
With the advent of a new sequencing platform, the genome of “CA” was re-assembled using 11.9 Gb long-read data from PacBio Sequel, and 94.2 Gb previously sequenced short-read data [23][7]. The new assembly of “CA” is 807.6 Mb with the contig N50 being 484.3 Kb, which has significantly increased the quality of the genome. The ratio of repetitive sequence (58.5%) was similar to the first version. Moreover, a cultivated lotus, “Taikonglian NO. 3”, was also assembled using the Oxford Nanopore sequencing platform (57.9 Gb raw data) with the contig N50 being 5.1 Mb, and eight chromosomes were anchored based on high-throughput chromatin conformation capture (Hi-C) data [24][8]. Another lotus species, American lotus, was recently assembled using PacBio RSII (74.6 Gb raw data) and Hi-C (50.32 Gb raw data), and the total length is 843 Mb while contig N50 is 1.34 Mb [25][9].
3. Potential Adaptive Evolution and Domestication of Lotus
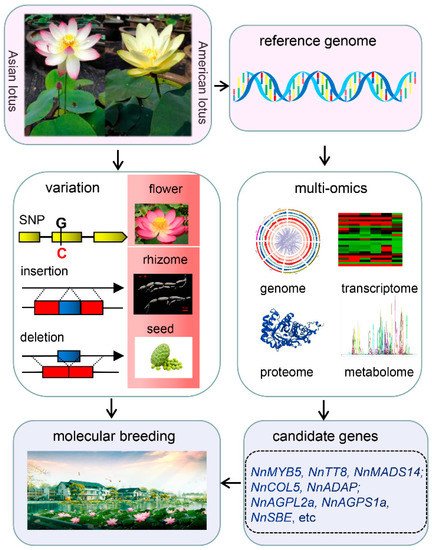
The availability of lotus reference genome information has facilitated the resequencing of different lotus germplasms. Several studies were conducted on how the lotus genome was subjected to adaptive evolution and artificial selection. Although it is known that there are only two species of lotus, namely Asian lotus and American lotus, except for the difference in flower color, their plant architecture and morphology are very similar. Based on molecular phylogeny analysis, significant genetic differentiation between American and Asian lotus was verified [25,26,27,28][9][10][11][12]. De-novo deep sequencing of the American lotus showed that its genome size is 843 Mb, and an approximate 81% repeat sequence was identified (Table 1), which is larger than the genome of Asian lotus. It is interesting to investigate the dramatic difference in repeat sequence between them because most protein-coding genes show a one-to-one synteny pattern. A total of 29,533 structure variations (SVs) were detected between two lotus species, with the SV-associated genes overexpressed in ‘regulation of mitotic cell cycle’, and ‘protein transporter activity’ [25][9]. Meanwhile, this study also showed that the selection on an MYB gene might contribute to the color difference between Asian and American lotus [25][9]. It is still an open question about when the two species diverged during the evolution and how they could keep high similarity in the independent geographical evolution. The wild lotus is distributed widely worldwide and maintains higher genomic diversity than cultivated lotus. Tropical and temperate lotus are the two ecotypes of Asian lotus. The comparison of the genome of these two ecotypes showed that a total of 453 genes were subjected to selection, including cyp714a genes that may relate to rhizome morphogenesis and a 10-Mb region in chromosome 1 that might play key roles in environmental adaption; including a homolog gene of at5g2394 in Arabidopsis encoding an acyltransferase protein [24][8]. By comparing their expressional patterns, the genes encoding granule-bound starch synthases, storage organ development, COSTAN-like gene family, vernalization, as well as cold response genes may relate to ecotypic differentiation [26][10]. It is very important to know the genetic backgrounds of parental lines in breeding. The origin, classification, and evolution of cultivated lotus were investigated through population re-sequence analysis. A total of 18 lotus accessions, including categories of American, seed, rhizome, flower, wild, and Thai lotus, were re-sequenced, based on which phylogenetic tree was constructed. The results indicated that the rhizome lotus had a closer relationship with wild lotus. In contrast, seed and flower lotus were admixed [26][10], which could be supported by re-sequencing of an enlarged population containing 296 accessions of different germplasm (58 wild, 163 rhizome, 39 flower, 32 seed lotus varieties) [28][12]. Further re-sequencing of 69 lotus accessions showed that flower lotus might mix with rhizome or seed lotus [27][11].4. Identification of Genes with Potential Application in Lotus Breeding
Flower color, shape, and flowering time are important traits that determine the ornamental value of lotus. There are three different colors in lotus, red and white in Asian lotus and yellow in American lotus. The red color in Asian lotus is determined by the contents of anthocyanin [40[13][14],41], which is controlled by key enzyme encoding genes, and their regulating transcription factors (TFs) such as MYB, basic-Helix-Loop-Helix (bHLH), WD40 in its biosynthetic pathway. Among all the enzyme encoding genes in this pathway, NnANS and NnUFGT seem to be the decisive two genes [42,43][15][16]. Several TFs including 5 MYB, 2 bHLH, and one WD-repeat genes, may be involved in the regulation of anthocyanin biosynthesis in lotus based on a transcriptome analysis [43][16]. Among them, a bHLH gene NnTT8 was verified to regulate anthocyanin biosynthesis [44][17], whereas the yellow color of American lotus is determined by carotenoid, and no anthocyanin was detected [25,45][9][18].
The rhizome is the main edible part of lotus. It is important to explore the mechanisms underlying rhizome formation and expansion in rhizome lotus breeding. Comparative transcriptomic and proteomic analyses focusing on rhizome development have been conducted to dig out the key genes and pathways critical for the crucial physiological process [52,53,54][19][20][21]. Furthermore, re-sequencing of the natural and genetic F2 populations has also identified several genetic regions and candidate genes that might be involved in lotus rhizome enlargement [55][22]. A systematic analysis was conducted on one candidate gene CONSTANS-LIKE 5 (COL5). Functional analysis in the potato system indicated that NnCOL5 might be positively associated with rhizome enlargement by regulating the expression of CO-FT genes and the GA signaling pathway [56][23]. In addition, one SNP was identified in another candidate gene NnADAP of AP2 subfamily, which is closely associated with rhizome enlargement phenotype and the soluble sugar content [57][24]. There is a big difference between temperate and tropical lotus, especially the rhizome’s morphology.
Lotus seeds are rich in nutrients and functional compounds such as alkaloids, flavonoids, and polyphenols [58,59][25][26]. They are consumed “as both food and medicine” [60][27]. It is essential to increase the yield and nutrition of lotus seed. The main factors determining lotus seed yield are the seed size and the number of lotus seeds per seedpod. Transcriptome analysis on the cotyledon of “CA” and “Jianxuan-17 (JX-17)” seeds at different developmental stages identified 8437 differentially expressed genes (DEGs). Many DEGs are involved in the brassinosteroid biosynthesis pathway, and further analysis predicted two AGPase genes as candidate genes affecting lotus seed yield [61][28]. It seems that phytohormones are involved in lotus seed development. A combination of metabolomic and proteomic methods revealed that 15 DAP (Day After Pollination) was a switch time point from the physiological active to the nutrition accumulation stage [62][29]. Starch is the primary nutritional component in mature lotus seed [63][30]. Its contents and the proportion of amylose and amylopectin could largely determine the nutritional value and taste of lotus cotyledon, respectively. ADP-glucose pyrophosphorylase (AGPase) plays an important role in regulating starch biosynthesis.
5. Conclusions
The new varieties of lotus with high yield, wide adaptability, and stress resistance play a vital role in improving the economic value of this important horticulture crop. The variations identification, functional gene cloning, and metabolites alterations among diverse germplasm resources were investigated in the past decades, driven by the progressively improved genome information which could facilitate breeding practices in lotus (Figure 1). However, a high-quality reference genome is the limiting factor that will affect the molecular breeding process. Improvement of the lotus reference genome will be a requisite in the future, directly affecting the accuracy of molecular markers and the efficiency of cloning functional genes. Gapless reference genomes and pan-genomes have become the new reference, based on which plentiful information of genomes such as open chromatin and more variant information can be explored. With the explosive growth of large-omics data, deep learning can be used to mine biological information and decipher gene regulation networks.
References
- Lin, Z.; Zhang, C.; Cao, D.; Damaris, R.N.; Yang, P. The latest studies on lotus (Nelumbo nucifera)-an emerging horticultural model plant. Int. J. Mol. Sci. 2019, 20, 3680.
- Shen-Miller, J. Sacred lotus, the long-living fruits of China Antique. Seed Sci. Res. 2002, 12, 131–143.
- Liu, L.; Li, Y.; Min, J.; Xiang, Y.; Tian, D. Analysis of the cultivar names and characteristics of global lotus (Nelumbo). Hans J. Agric. Sci. 2019, 9, 163–181.
- Ming, R.; VanBuren, R.; Liu, Y.; Yang, M.; Han, Y.; Li, L.T.; Zhang, Q.; Kim, M.J.; Schatz, M.C.; Campbell, M.; et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 2013, 14, R41.
- Wang, Y.; Fan, G.; Liu, Y.; Sun, F.; Shi, C.; Liu, X.; Peng, J.; Chen, W.; Huang, X.; Cheng, S.; et al. The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 2013, 76, 557–567.
- Gui, S.; Peng, J.; Wang, X.; Wu, Z.; Cao, R.; Salse, J.; Zhang, H.; Zhu, Z.; Xia, Q.; Quan, Z.; et al. Improving Nelumbo nucifera genome assemblies using high-resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 2018, 94, 721–734.
- Shi, T.; Rahmani, R.S.; Gugger, P.F.; Wang, M.; Li, H.; Zhang, Y.; Li, Z.; Wang, Q.; Van de Peer, Y.; Marchal, K.; et al. Distinct expression and methylation patterns for genes with different fates following a single whole-genome duplication in flowering plants. Mol. Biol. Evol. 2020, 37, 2394–2413.
- Zheng, X.; Wang, T.; Cheng, T.; Zhao, L.; Zheng, X.; Zhu, F.; Dong, C.; Xu, J.; Xie, K.; Hu, Z.; et al. Genomic variation reveals demographic history and biological adaptation of the ancient relictual, lotus (Nelumbo Adans). Hortic. Res. 2022, 9, uhac029.
- Zheng, P.; Sun, H.; Liu, J.; Lin, J.; Zhang, X.; Qin, Y.; Zhang, W.; Xu, X.; Deng, X.; Yang, D.; et al. Comparative analyses of American and Asian lotus genomes reveal insights into petal color, carpel thermogenesis and domestication. Plant J. 2022, 110, 1498–1515.
- Huang, L.; Yang, M.; Li, L.; Li, H.; Yang, D.; Shi, T.; Yang, P. Whole genome re-sequencing reveals evolutionary patterns of sacred lotus (Nelumbo nucifera). J. Integr. Plant Biol. 2018, 60, 2–15.
- Li, Y.; Zhu, F.L.; Zheng, X.W.; Hu, M.L.; Dong, C.; Diao, Y.; Wang, Y.W.; Xie, K.Q.; Hu, Z.L. Comparative population genomics reveals genetic divergence and selection in lotus, Nelumbo nucifera. BMC Genom. 2020, 21, 146.
- Liu, Z.; Zhu, H.; Zhou, J.; Jiang, S.; Wang, Y.; Kuang, J.; Ji, Q.; Peng, J.; Wang, J.; Gao, L.; et al. Resequencing of 296 cultivated and wild lotus accessions unravels its evolution and breeding history. Plant J. 2020, 104, 1673–1684.
- Yang, R.Z.; Wei, X.L.; Gao, F.F.; Wang, L.S.; Zhang, H.J.; Xu, Y.J.; Li, C.H.; Ge, Y.X.; Zhang, J.J.; Zhang, J. Simultaneous analysis of anthocyanins and flavonols in petals of lotus (Nelumbo) cultivars by high-performance liquid chromatography-photodiode array detection/electrospray ionization mass spectrometry. J. Chromatogr. A 2009, 1216, 106–112.
- Deng, J.; Chen, S.; Yin, X.; Wang, K.; Liu, Y.; Li, S.; Yang, P. Systematic qualitative and quantitative assessment of anthocyanins, flavones and flavonols in the petals of 108 lotus (Nelumbo nucifera) cultivars. Food Chem. 2013, 139, 307–312.
- Deng, J.; Fu, Z.; Chen, S.; Damaris, R.N.; Wang, K.; Li, T.; Yang, P. Proteomic and epigenetic analyses of lotus (Nelumbo nucifera) petals between red and white cultivars. Plant Cell Physiol. 2015, 56, 1546–1555.
- Deng, J.; Li, J.; Su, M.; Chen, L.; Yang, P. Characterization of key genes involved in anthocyanins biosynthesis in Nelumbo nucifera through RNA-Seq. Aquat. Bot. 2021, 21, 103428.
- Deng, J.; Li, J.; Su, M.; Lin, Z.; Chen, L.; Yang, P. A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis. Plant Physiol. Biochem. 2021, 158, 518–523.
- Sun, S.; Gugger, P.; Wang, Q.; Chen, J. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ 2016, 4, e2369.
- Cheng, L.; Li, S.; Yin, J.; Li, L.; Chen, X. Genome-wide analysis of differentially expressed genes relevant to rhizome formation in lotus root (Nelumbo nucifera Gaertn). PLoS ONE 2013, 8, e67116.
- Yang, M.; Zhu, L.; Pan, C.; Xu, L.; Liu, Y.; Ke, W.; Yang, P. Transcriptomic analysis of the regulation of rhizome formation in temperate and tropical lotus (Nelumbo nucifera). Sci. Rep. 2015, 5, 13059.
- Cao, D.; Damaris, R.N.; Zhang, Y.; Liu, M.; Li, M.; Yang, P. Proteomic analysis showing the signaling pathways involved in the rhizome enlargement process in Nelumbo nucifera. BMC Genom. 2019, 20, 766.
- Huang, L.; Li, M.; Cao, D.; Yang, P. Genetic dissection of rhizome yield-related traits in Nelumbo nucifera through genetic linkage map construction and QTL mapping. Plant Physiol. Biochem. 2021, 160, 155–165.
- Cao, D.; Lin, Z.; Huang, L.; Damaris, R.N.; Li, M.; Yang, P. A CONSTANS-LIKE gene of Nelumbo nucifera could promote potato tuberization. Planta 2021, 253, 65.
- Cao, D.; Lin, Z.; Huang, L.; Damaris, R.N.; Yang, P. Genome-wide analysis of AP2/ERF superfamily in lotus (Nelumbo nucifera) and the association between NnADAP and rhizome morphology. BMC Genom. 2021, 22, 171.
- Limwachiranon, J.; Huang, H.; Shi, Z.; Li, L.; Luo, Z. Lotus flavonoids and phenolic acids, health promotion and safe consumption dosages. Compr. Rev. Food Sci. Food Saf. 2018, 17, 458–471.
- Yu, Y.; Wei, X.; Liu, Y.; Dong, G.; Hao, C.; Zhang, J.; Jiang, J.; Cheng, J.; Liu, A.; Chen, S. Identification and quantification of oligomeric proanthocyanidins, alkaloids, and flavonoids in lotus seeds, A potentially rich source of bioactive compounds. Food Chem. 2022, 379, 132124.
- Zhang, Y.; Lu, X.; Zeng, S.; Huang, X.; Guo, Z.; Zheng, Y.; Tian, Y.; Zheng, B. Nutritional composition, physiological functions and processing of lotus (Nelumbo nucifera gaertn.) seeds, a review. Phytochem. Rev. 2015, 14, 321–334.
- Li, J.; Tao, S.; Huang, L.; He, D.; Maraga, N.T.; Yang, P. Systematic transcriptomic analysis provides insights into lotus (Nelumbo nucifera) seed development. Plant Growth Regul. 2018, 86, 339–350.
- Wang, L.; Fu, J.; Li, M.; Fragner, L.; Weckwerth, W.; Yang, P. Metabolomic and proteomic profiles reveal the dynamics of primary metabolism during seed development of lotus (Nelumbo nucifera). Front. Plant Sci. 2016, 7, 750.
- Shen-Miller, J.; Mudgett, M.B.; Schopf, J.W.; Clarke, S.; Berger, R. Exceptional seed longevity and robust growth, ancient sacred lotus from china. Am. J. Bot. 1995, 82, 1367–1380.
