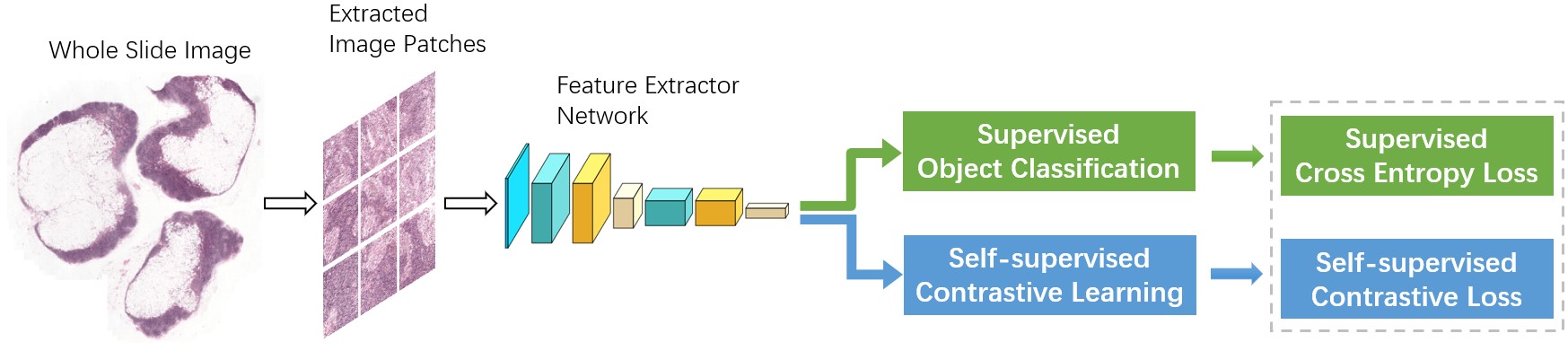
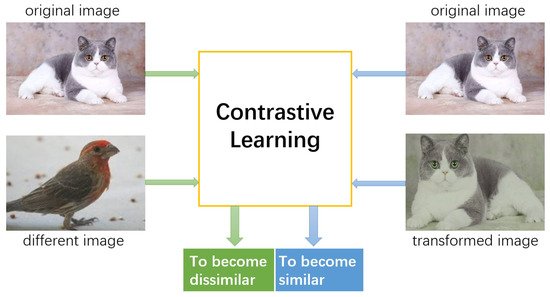
The metastasis detection in lymph nodes via microscopic examination of H&E stained histopathological images is one of the most crucial diagnostic procedures for breast cancer staging. The manual analysis is extremely labor-intensive and time-consuming because of complexities and diversities of histopathological images. Deep learning has been utilized in automatic cancer metastasis detection in recent years. The success of supervised deep learning is credited to a large labeled dataset, which is hard to obtain in medical image analysis. Contrastive learning, a branch of self-supervised learning, can help in this aspect through introducing an advanced strategy to learn discriminative feature representations from unlabeled images. In this paper, we propose to improve breast cancer metastasis detection through self-supervised contrastive learning, which is used as an accessional task in the detection pipeline, allowing a feature extractor to learn more valuable representations, even if there are fewer annotation images. Furthermore, we extend the proposed approach to exploit unlabeled images in a semi-supervised manner, as self-supervision does not need labeled data at all. Extensive experiments on the benchmark Camelyon2016 Grand Challenge dataset demonstrate that self-supervision can improve cancer metastasis detection performance leading to state-of-the-art results.
- convolutional neural network
- contrastive learning
- self-supervision
- deep learning
1. Introduction
2. RBrelated Workast Cancer Detection
In this section, we provide an overview of the relevant literature on breast cancer detection and self-supervised contrastive learning.
2.1 Breast Cancer Detection
2.2 Self-Supervised Contrastive Learning
3. Self-Supervised Contrastive Learning
3. Methodology
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Ramos-Vara, J.A. Principles and methods of immunohistochemistry. Methods Mol. Biol. 2011, 691, 83–96.
- Humphreys, G.; Ghent, A. World laments loss of pathology service. Bull. World Health Organ. 2010, 88, 564–565.
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2015, 63, 1455–1462.
- Cruz-Roa, A.A.; Ovalle, J.; Madabhushi, A.; Osorio, F. A Deep Learning Architecture for Image Representation, Visual Interpretability and Automated Basal-Cell Carcinoma Cancer Detection. In Proceedings of the 16th International Conference on Medical Image Computing and Computer Assisted Intervention, Nagoya, Japan, 22–26 September 2013; pp. 403–410.
- Kandemir, M.; Hamprecht, F.A. Computer-aided diagnosis from weak supervision: A benchmarking study. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2015, 42, 44–50.
- Spanhol, F.; Oliveira, L.S.; Cavalin, P.R.; Petitjean, C.; Heutte, L. Deep features for breast cancer histopathological image classification. In Proceedings of the IEEE International Conference on Systems, Man, and Cybernetics, Banff, AB, Canada, 5–8 October 2017; pp. 1868–1873.
- Bayramoglu, N.; Kannala, J.; Heikkilä, J. Deep learning for magnification independent breast cancer histopathology image classification. In Proceedings of the 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 2440–2445.
- Guo, Y.; Dong, H.; Song, F.; Zhu, C.; Liu, J. Breast Cancer Histology Image Classification Based on Deep Neural Networks. In International Conference Image Analysis and Recognition; Springer: Cham, Switzerland, 2018; Volume 10882, pp. 827–836.
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Essen, B.C.V.; Awwal, A.A.S.; Asari, V.K. The History Began from AlexNet: A Comprehensive Survey on Deep Learning Approaches. arXiv 2018, arXiv:1803.01164.
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88.
- Ehteshami Bejnordi, B.; Linz, J.; Glass, B.; Mullooly, M.; Gierach, G.; Sherman, M.; Karssemeijer, N.; van der Laak, J.; Beck, A. Deep learning-based assessment of tumor-associated stroma for diagnosing breast cancer in histopathology images. In Proceedings of the IEEE 14th International Symposium on Biomedical Imaging, Melbourne, VIC, Australia, 18–21 April 2017; pp. 929–932.
- Lin, H.; Chen, H.; Dou, Q.; Wang, L.; Qin, J.; Heng, P.A. ScanNet: A Fast and Dense Scanning Framework for Metastatic Breast Cancer Detection from Whole-Slide Images. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 539–546.
- Lin, H.; Chen, H.; Graham, S.; Dou, Q.; Rajpoot, N.; Heng, P.A. Fast ScanNet: Fast and Dense Analysis of Multi-Gigapixel Whole-Slide Images for Cancer Metastasis Detection. IEEE Trans. Med. Imaging 2019, 38, 1948–1958.
- Zanjani, F.G.; Zinger, S.; With, P. Cancer detection in histopathology whole-slide images using conditional random fields on deep embedded spaces. In Proceedings of the Digital Pathology, Houston, TX, USA, 6 March 2018.
- Yi, L.; Wei, P. Cancer Metastasis Detection with Neural Conditional Random Field. arXiv 2018, arXiv:1806.07064.
- Kong, B.; Xin, W.; Li, Z.; Qi, S.; Zhang, S. Cancer Metastasis Detection via Spatially Structured Deep Network. In International Conference Image Analysis and Recognition; Springer: Cham, Switzerland, 2017; pp. 236–248.
- Xie, J.; Liu, R.; Luttrell, J.; Zhang, C. Deep Learning Based Analysis of Histopathological Images of Breast Cancer. Front. Genet. 2019, 10, 80.
- de Matos, J.; de Souza Britto, A.; Oliveira, L.; Koerich, A.L. Double Transfer Learning for Breast Cancer Histopathologic Image Classification. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8.
- Kassani, S.H.; Kassani, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Breast Cancer Diagnosis with Transfer Learning and Global Pooling. In Proceedings of the International Conference on Information and Communication Technology Convergence (ICTC), Jeju, Korea, 16–18 October 2019; pp. 519–524.
- Doersch, C.; Gupta, A.; Efros, A.A. Unsupervised Visual Representation Learning by Context Prediction. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1422–1430.
- Pathak, D.; Krähenbühl, P.; Donahue, J.; Darrell, T.; Efros, A.A. Context Encoders: Feature Learning by Inpainting. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2536–2544.
- Noroozi, M.; Favaro, P. Unsupervised Learning of Visual Representations by Solving Jigsaw Puzzles. In Proceedings of the ECCV, Amsterdam, The Netherlands, 11–14 October 2016.
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised Representation Learning by Predicting Image Rotations. arXiv 2018, arXiv:1803.07728.
- Zhang, R.; Isola, P.; Efros, A.A. Colorful Image Colorization. In Proceedings of the ECCV, Amsterdam, The Netherlands, 11–14 October 2016.
- Chen, T.; Zhai, X.; Ritter, M.; Lucic, M.; Houlsby, N. Self-Supervised GANs via Auxiliary Rotation Loss. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 12146–12155.
- Kolesnikov, A.; Zhai, X.; Beyer, L. Revisiting Self-Supervised Visual Representation Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 1920–1929.
- Hadsell, R.; Chopra, S.; LeCun, Y. Dimensionality Reduction by Learning an Invariant Mapping. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, New York, NY, USA, 17–22 June 2006; Volume 2, pp. 1735–1742.
- Wu, Z.; Xiong, Y.; Yu, S.X.; Lin, D. Unsupervised Feature Learning via Non-parametric Instance Discrimination. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3733–3742.
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R.B. Momentum Contrast for Unsupervised Visual Representation Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 9726–9735.
- Misra, I.; van der Maaten, L. Self-Supervised Learning of Pretext-Invariant Representations. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 6706–6716.
- Tian, Y.; Krishnan, D.; Isola, P. Contrastive Multiview Coding. In Proceedings of the ECCV, Glasgow, UK, 23–28 August 2020.
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G.E. A Simple Framework for Contrastive Learning of Visual Representations. arXiv 2020, arXiv:2002.05709.
- Dosovitskiy, A.; Springenberg, J.T.; Riedmiller, M.; Brox, T. Discriminative Unsupervised Feature Learning with Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems 27 (NIPS), Montreal, QC, Canada, 8–13 December 2014.
- Tschannen, M.; Djolonga, J.; Ritter, M.; Mahendran, A.; Houlsby, N.; Gelly, S.; Lucic, M. Self-Supervised Learning of Video-Induced Visual Invariances. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 13803–13812.
- Bachman, P.; Hjelm, R.D.; Buchwalter, W. Learning Representations by Maximizing Mutual Information Across Views. In Proceedings of the NeurIPS, Vancouver, BC, Canada, 8–14 December 2019.
- Hénaff, O.J.; Srinivas, A.; Fauw, J.D.; Razavi, A.; Doersch, C.; Eslami, S.M.A.; van den Oord, A. Data-Efficient Image Recognition with Contrastive Predictive Coding. arXiv 2020, arXiv:1905.09272.
- Hjelm, R.D.; Fedorov, A.; Lavoie-Marchildon, S.; Grewal, K.; Trischler, A.; Bengio, Y. Learning deep representations by mutual information estimation and maximization. arXiv 2019, arXiv:1808.06670.
- Tschannen, M.; Djolonga, J.; Rubenstein, P.K.; Gelly, S.; Lucic, M. On Mutual Information Maximization for Representation Learning. arXiv 2019, arXiv:1907.13625.
- Caron, M.; Misra, I.; Mairal, J.; Goyal, P.; Bojanowski, P.; Joulin, A. Unsupervised Learning of Visual Features by Contrasting Cluster Assignments. arXiv 2020, arXiv:2006.09882.
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, F.F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Miami, FL, USA, 20–25 June 2009.
