Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 3 by Sirius Huang.
Electrochemical biosensors have tremendous potential for point-of-care virus detection due to their portability and simple detection methods. The main limitation of this method is its sensitivity. Numerous studies have been conducted in an attempt to enhance the key elements of electrochemical-based biosensing systems.
- electrochemical biosensors
- virus detection
- potentiostat
- cyclic voltammetry
1. Introduction
An electrochemical biosensor plays three crucial roles: biorecognition, transduction and signal output, as shown in Figure 1 [1].
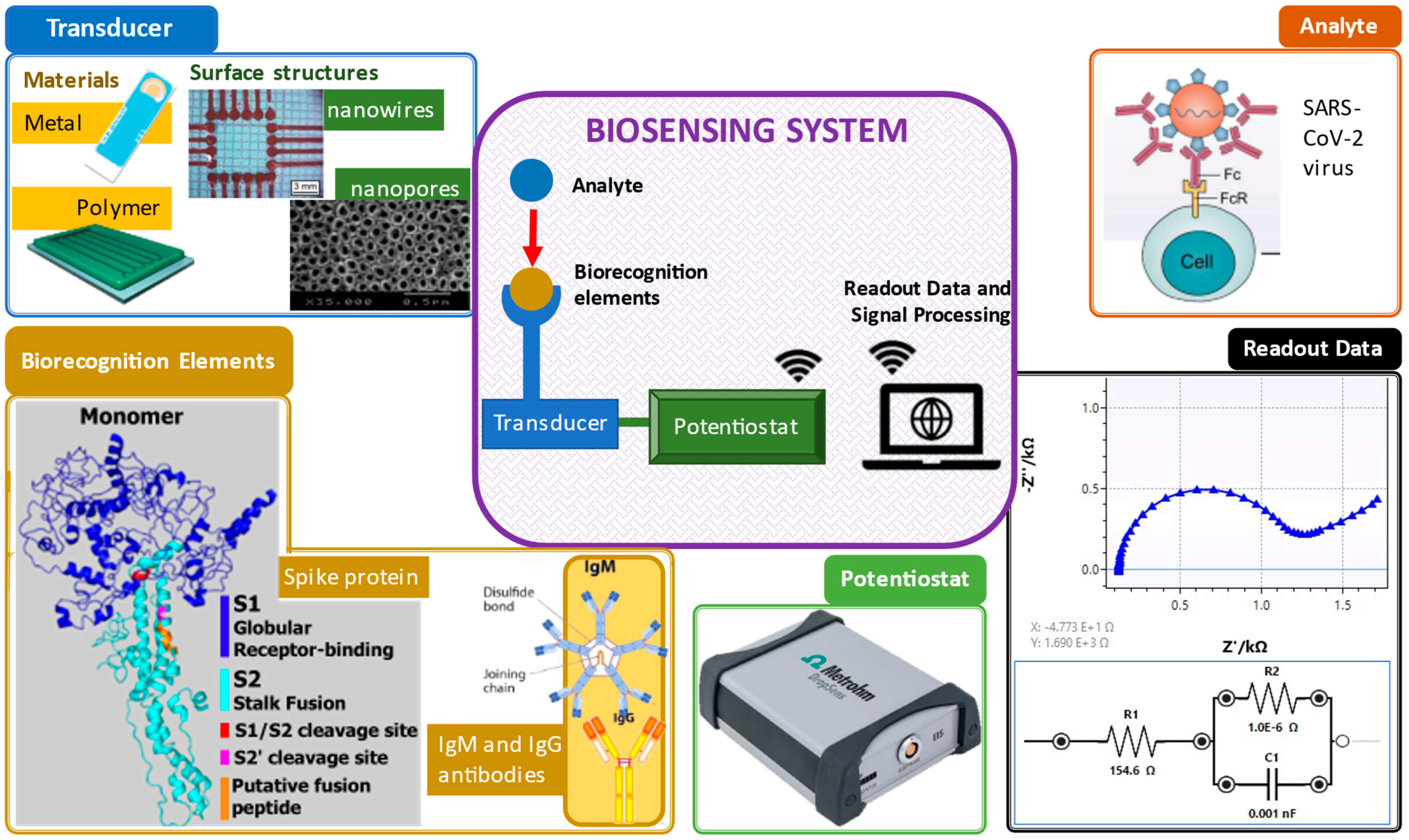
Figure 1. Important elements in a biosensing system. Clockwise from top right: analyte [2], electrochemical impedance spectroscopy readout data, potentiostat [3], biorecognition elements (spike protein [4] and antibodies [5]) and transducers and their surface structures (screen-printed gold electrode [6], polymer electrode [7], nanowire structure [8] and nanopore structure [9]).
In general, the principle of biosensors involves, first, the identification of biorecognition to the target samples. Next, the transducer or electrodes detect any changes due to the interaction of both the target and biorecognition. Then, the transducer translates the interaction changes to a digital detector. Lastly, the digital output signals are displayed by digital devices, such as smartphones or laptops [10]. The digital output signals can be described as the electrochemical response, including the increase (signal on) and decrease (signal off) of the target concentration elevation by the electrodes measured via either electron transfer or electron transfer resistance [11]. With the existing biorecognition elements mixed with the analyte, existence of virus detected in the sample can be confirmed.
2. Theory of Electrochemical Biosensors
Electrochemistry is one of the key measuring methods to view reactions involved in electron transfers [12]. Electrochemical biosensors usually contain biological recognition elements that react selectively with the target analytes. Biological recognition elements are typically enzymes, antibodies or nucleic acid [13] that react to the tested analytes, such as viruses, as shown in Figure 1. Electrochemical biosensors can be divided into two categories: biocatalytic sensors and affinity biosensors. Biocatalytic sensors use enzymes as recognition elements and are commonly used for blood glucose monitoring, whereas affinity sensors use biomolecules, such as antibodies, oligonucleotides or commonly used aptamers, as biorecognition elements [14]. Affinity biosensors typically utilize antibodies or aptamers for virus detection. Figure 2 shows the important components that are necessary for successful virus detection.
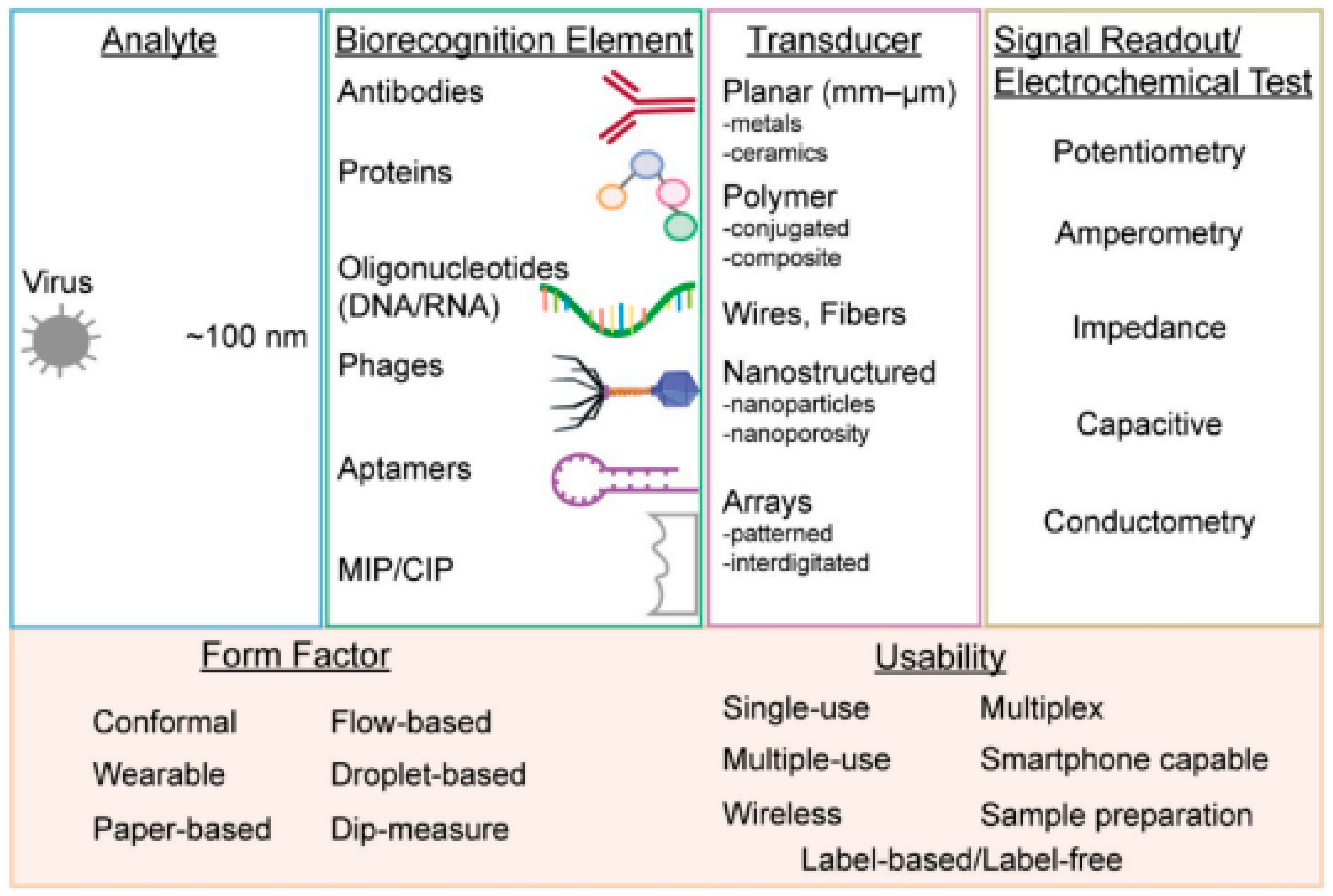
Figure 2. From left to right: virus form factors, types of biorecognition elements, methods to enhance the sensitivity of transducers, various measurement techniques using electrochemical testing and the multi-usability of electrochemical biosensors [14].
Various electrochemical techniques that can be used for biosensing, such as voltammetry, impedance spectroscopy, amperometry, chronopotentiometry, etc. Voltammetry measures current when different potentials are applied. By performing voltammetry, voltage and current characteristics of a medium can be determined, as well as mechanistic and kinetic details of electron transfer [15]. Cyclic voltammetry (CV) is usually used to investigate the reduction process of molecular species, as well as oxidation processes, making it important to study the chemical reactions that involve electron transfer and can also be used to characterize the stability of electrodes [12]. Electrochemical impedance spectroscopy (EIS) is a technique used to investigate electrical properties of various forms of matter by measuring the current response after applying sinusoidal voltage [16]. According to the current response, the data can be represented in three forms (Fourier transform) in order to decompose the result to assess the magnitude and frequency in the form of a bode plot to measure the frequency response of systems, in addition to a Nyquist plot. The results from the Nyquist plot can determine double-layer capacitance, the ohmic resistance of the electrolyte solution, Warburg impedance and electron transfer resistance. EIS is also a powerful technique that can be used to analyze the electron transfer kinetics and for the detection of molecular interactions.
These three electrochemical techniques each have their advantages and disadvantages depending on how they are executed. With respect to time taken for execution, amperometry and voltammetry techniques can be applied with a shorter duration than EIS. The execution time usually depends on the scan rate [17], which impacts the time taken for the operation to complete, which can be as short as 10 s or as long as a few minutes. However, electrochemical impedance spectroscopy (EIS) techniques take much longer. Normally, EIS is executed in the frequency range of 0.1Hz to 100KHz. The time taken for EIS to be executed in this range usually depends on the microcontroller performances of the potentiostat. For example, EIS running on μStat-i 400 from MetroOhm with the stated frequency range and 10 readings per decade takes approximately 10-15 min, whereas with the Pico development kit from Palmsens, the same operations takes approximately 27 min.
Impedance characterization of the electrode–electrolyte interface is important in the field of impedance-based biosensing. EIS is a useful technique, as it captures the phenomena that occur at the electrode interface. Impedance is measured by applying a sinusoidal voltage to the electrochemical cell and measuring the produced current. The mathematical basis of the system can be explained by Equation (1) [18].
where Z is impedance, and both Et and It are potential and current at a specific time instant. A phase shift occurs between the applied voltage and the current due to the capacitive and resistive effects of the electrochemical system [19]. The electrode–electrolyte interface includes a charge transfer resistance and capacitance in combination with the resistance of the solution resistance. This impedance data over a set frequency range can be informative with respect to any ionic interactions or modifications that occur on the surface of the electrode. Equivalent circuit models, such as Warburg circuits, can be used to model these interactions and can be curve-fitted to the measured data. The selected model of the equivalent circuit is deemed acceptable if it has low chi-squared values (χ2) [20].
Z = Et/It (1)
In terms of measurement durations, amperometry and voltammetry are far superior to EIS. However, timing performance is not the only indicators that needs to be taken into consideration. Amperometry and voltammetry techniques can be used to detect electron transfer through oxidation/reduction reaction by supplying currents or potentials [21]. EIS has advantages over amperometry in terms of direct antibody–antigen binding detection without applying high potentials to induce a reduction/oxidation reaction [21]. Therefore, all the mentioned advantages and disadvantages should be considered selecting the optimal techniques for detection.
3. Important Design Considerations for Detection of Viruses
One important consideration for detection of viruses is that the size of a virus is smaller than that of bacteria [22]. Most viruses have sizes ranging from 10 to 100 nm, compared to bacteria, which are at least 10 times smaller, given that the typical bacterium is approximately 1 µm in size [23]. Because sizes are in the nanometer range, other than sensitivity and selective biorecognition elements, detection of viruses often requires amplification, immobilization of sensing materials and surface passivation to enhance binding reactions [14].
3.1. Transducing Elements
A key component of a biosensor is the transducer, which can transform chemical interactions on the sensing surface into electrical signals via electrodes. Typically, a three-electrode format is used in electrochemical systems. Electrodes can be fabricated from conducting and semi-conducting materials, such as carbon, which is widely used, as well as gold (Au). Electrode designs can vary in terms of form factors, usability, materials as and fabrication method. Integrated electrodes can be manufactured cost-effectively using techniques such as screen printing and lamination methods. Screen-printed electrodes (SPEs) are commercially manufactured and can produce stable, reliable results, making them ideal as a cost-effective solution for high-throughput screening that requires disposable devices. Polymer electrodes have also been used for virus detection. One example is a graphene–polymer electrode used for detection of dengue virus (DENV) [24]. The use of polymers as electrode elements is associated with various advantages, such as tunable electrical conductivity, biocompatibility and environmental stability. Electrodes are also compatible with multiple ranges of immobilization techniques and biorecognition elements [25][26].
In order to better understand how electrode materials impact detection, an experiment was conducted to compare the performance of commercialized carbon and gold electrodes. Carbon has a conductivity of 1.25 × 103 to 2.00 × 103 S/m at 20 °C, whereas gold has a much higher conductivity of 4.11 × 107 S/m at 20 °C. As shown in Figure 3, SPGE shows higher oxidation and reduction peaks compared to SPCE due to its higher conductivity. Despite its superior performance, SPGE is less commonly used compared to SPCE due to its higher associated costs. Thus, a considerable amount of research has been conducted to tackle this issue in an attempt improve the sensitivity of SPCE, mainly by increasing the electrode surface area. The larger the surface area, the higher the sensitivity of the electrode, which is particularly crucial for virus detection [27][28].
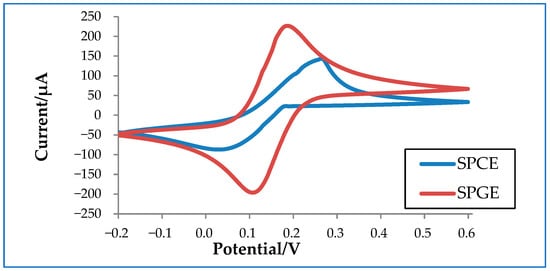
Figure 3. Cyclic voltammetry graph of a commercialized screen-printed carbon electrode (SPCE) and a screen-printed gold electrode (SPGE) in 10 mM ferrocyanide.
Higher surface area can be achieved by nanostructure formations on top of the electrodes. Examples of these nanostructures are nanowires and nanopores [29]. Molecular imprinted polymers (MIPs) can also be used to enhance the surface area and result in improved electrode performance, as shown in Figure 4 [7]. In this work, MIPs were electropolymerized on commercial carbon electrodes to produce an enhanced cyclic voltammetry response for detection of SARS-CoV-2 viruses. The peak separation and current values are much higher in electrodes with a nanoporous gold thin film compared to bare gold electrodes.
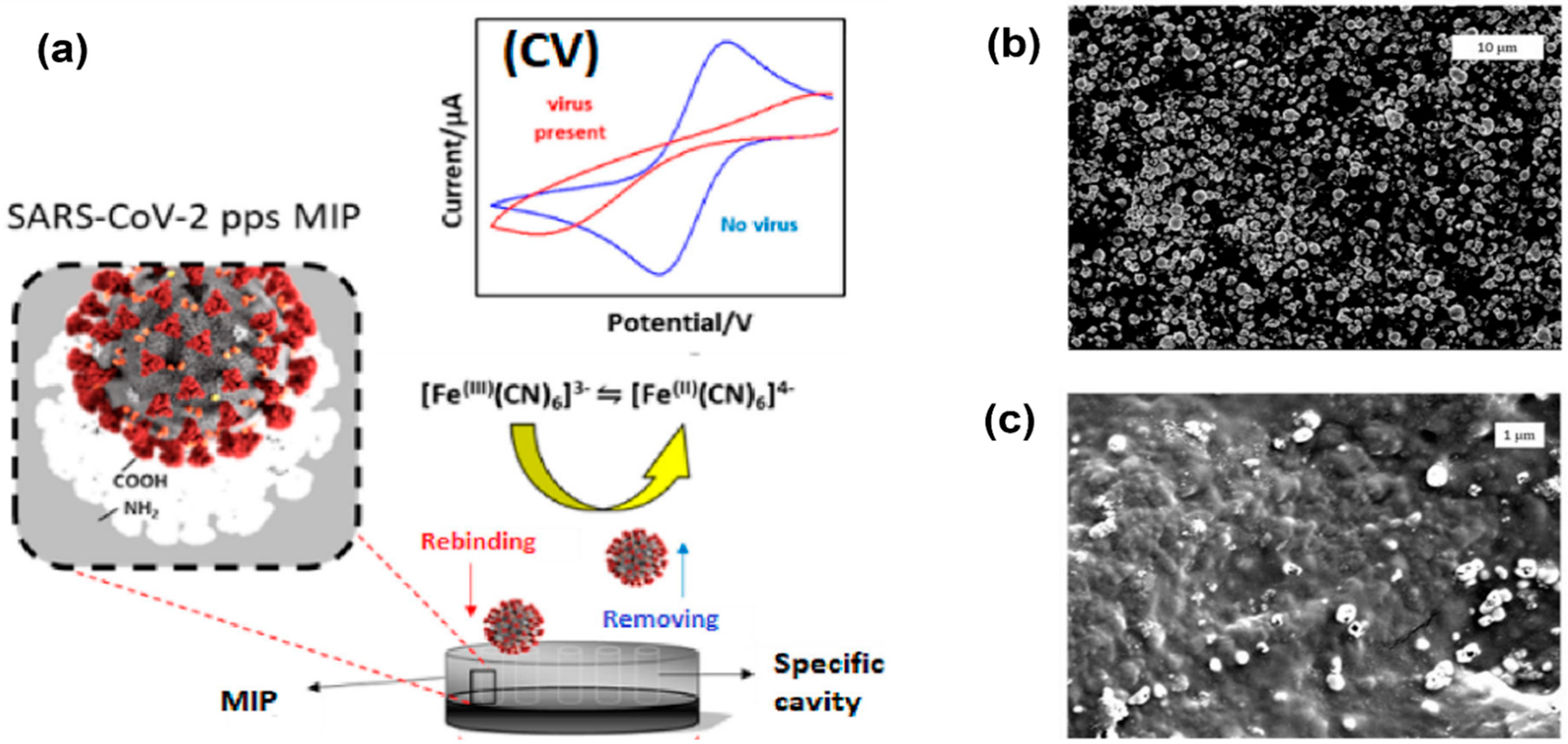
Figure 4. Electropolymerization technique using molecular imprinted polymer (MIP) to enhance the surface area and sensitivity of electrodes. (a) Imprinted MIP is electrodeposited on a screen-printed carbon electrode to create enhanced anodic/cathodic currents during cyclic voltammetry measurements. (b) Electrode surface before polymerization. (c) Electrode surface after polymerization, indicating polymer modification and a change in topography [30].
Figure 5 shows the cyclic voltammetry graph of bare gold, nanoporous gold thin film (NPGF), NPGF with hemoglobin immobilized with 6-mercaptohexanoic acid (6-MHA) and NPGF with hemoglobin immobilized with DNA. The bare gold electrode and the NPGF oxidation and reduction peaks prove that with increasing surface area of the electrode, the detection performance improves.
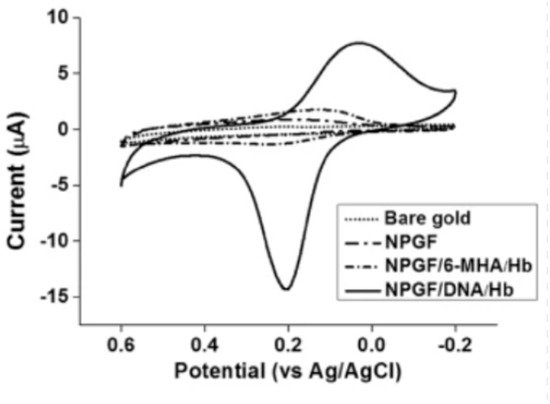
Figure 5. Cyclic voltammograms of bare gold, a nanoporous gold thin film (NPGF) electrode, NPGF/6-MHA/Hb and NPGF/DNA/Hb in PBS [31].
Several studies have been conducted in an effort to improve the surface area of electrodes by adding various types of nanostructures. Cost efficiency can also be improved by coating low-cost electrodes with high-conductivity materials with nanostructures, for example, gold-nanoparticle-coated, screen-printed carbon electrodes [32].
3.2. Biorecognition Elements
Biorecognition elements are the sensing materials that are immobilized on the surface of a biosensor platform. Biorecognition material refers to nucleic acid, antibodies, enzymes and cell receptors that are mounted over a transducer and react with the target analyte in the solution to generate a biochemical response [33]. The bioreceptors identify and bind the target while minimizing any interference from other compounds and organisms with high selectivity and sensitivity [34]. The interface between the analyte and the bioreceptor is subsequently translated by the transducer and emits biological signals [35]. Biorecognition elements can be used from naturally occurring states or from synthetic constructs. These elements can be used, along with transducers, before performing electrochemical techniques.
Common biorecognition elements for virus detection include antibodies and antibody fragments. Antibodies have recognition sites that selectively bind to antigens via a specific region of the antigen known as an epitope [36]. Antibodies can be labelled, or fluorescent or enzymatic tags can be attached. This includes additional reagents and processing steps; however, the binding affinity to the antigen can influence the biosensor’s selectivity [37][38]. Biosensors employing antibody-based biorecognition elements are commonly referred to as immunosensors. Given that antibodies exhibit high selectivity and binding affinity for target species and can be generated for a wide range of infectious agents, they are the gold-standard biorecognition element for virus detection.
Protein detection of COVID-19 can be classified into three components: detection of nucleocapsids, spike proteins and/or a combination of both nucleocapsids and spike proteins [39]. An example of an electrochemical biosensor that detects nucleocapsids is a graphene-based biosensor developed by Alafeef et al. with an integrated electrical readout [40]. This biosensor employs thiol-modified antisense oligonucleotides probes (ssDNA probes) with capped gold nanoparticles (AuNPs) that are specific and selective to detect the nucleocapsid (N-gene) of COVID-19. For detection of spike protein (S protein), an electrochemical sensor with cobalt-functionalized TiO2 nanotubes (Co-TNTs) was developed by Vadlamani et al. [41]. The Co-TNTs act as a sensing material to detect the S-protein receptor binding domain of COVID-19. An example of an electrochemical biosensor that detects both nucleocapsids and spike glycoproteins is multiplex rolling circle amplification (RCA) [42]. RCA is an isothermal amplification technique involving DNA or RNA primers annealed to a circular DNA template with the help of DNA or RNA polymerase. An RCA amplicon contains multiple copies of DNA sequences in an end-to-end tandem arrangement known as a concatemer corresponding to a circular template. This type of electrochemical biosensor can rapidly detect S and N genes of COVID-19 in clinical samples, including sandwich hybridization of RCA amplicons with probes and redox active labels known as silica-methylene blue (SiMB) and silica-acridine orange (SiAO). The signal output of the device is detected by differential pulse voltammetry (DPV) [42]. Another example of a COVID-19 sensor using antibodies is a label-free, paper-based electrochemical platform [43]. This type of sensor detects IgM and IgG antibodies that develop in the human body in response to the COVID-19 infection. An immunocomplex is formed between captured antibodies with the immobilized spike protein of COVID-19 antigen on the surface of the electrodes, causing a decrease in current. The current response to the formation of immunocomplex is recorded using the square-wave voltammetry (SWV) method.
Other than antibodies, another type of probe that is gaining traction in the field of biosensors is aptamers. Aptamers are single-stranded oligonucleotides with the ability to bind to various molecules with high selectivity and affinity [44]. Aptamers are isolated from a large random sequence pool through SELEX, which is a process of selection involving systematic evolution of ligands by exponential enrichment [45]. Suitable binding sequences can be isolated from oligonucleotide sequence pools before being amplified for usage; thus, the aptamers can exhibit high selectivity to target species [45]. Aptamers are also known as “chemical antibodies” and show benefits relative to antibodies, such as quick synthesis and excellent stability. Aptamers can also be fabricated at a lower cost than other biorecognition elements, such as antibodies. Aptamers have been used extensively in fundamental research, sensing and even as medications to cure illnesses and stop the spread of viruses [46][47].
Pathogen and virus detection using electrochemical biosensors usually employ single-stranded DNA aptamers. Electrochemical aptasensors can be classified into aptasensors without enzymes and aptasensors with enzymes. Aptasensors without enzymes involve the deposition of aptamers on the electrode. Binding between aptamers and the target causes direct changes in impedance. An example of this type of aptasensor is a sensor for detection of avian influenza A (AIV H5N1) [48]. This impedance aptasensor has a select DNA aptamer specific to AIV H5N1 placed on the gold microelectrode surface housed in a microfluidics flow cell, with a detection limit of 0.0128 hemagglutinin units (HAU) and a detection time of 30 min. The DNA aptamer is specific to H5N1 and does not react with other AIV subtypes, such as H1N1, H2N2 or H7N2.
The next type of electrochemical aptasensor is aptasensors with enzymes, whereby enzyme catalysis aids in the modulation of the electrical signal. An example of this type of aptasensor was developed by Ganabri et al. for the detection of hepatitis C virus (HCV) [49]. The aptamer specific to the HCV core antigen is embedded on graphene quantum dots (GQD) on the surface of a glassy carbon electrode (GCE). This aptasensor has a detection limit of 3.3 pg/mL, with two separate linearity ranges: 10–70 pg/mL and 70–400 pg/mL [49].
Another technique that is rapidly gaining popularity is molecular imprinting technology (MIT) which is an artificial approach to designing strong recognition materials that can resemble natural recognition materials, such as antibodies, and biologically and chemically certify molecules such as amino acids, proteins, nucleotides derivatives, etc. [50][51]. Using MIT, complex linkages between analytes and functional monomers are formed, called molecular imprinted polymers (MIPs), providing a recognition site corresponding to the shape of and aligned to the target molecules [52]. MIPs are porous substances that have been effectively used as synthetic receptors for a wide range of targets, including viruses, biomarkers and explosives. They feature high-affinity binding sites for each target molecule [53]. There are various types of MIP-based sensors; the content herein focuses on examples of electrochemical MIP-based sensors.
An MIP-based impedimetric sensor was recently developed for the detection of dengue infection in an early stage [54]. For this sensor, non-structural protein 1 (NS1) was used as a template during polymerization on a screen-printed carbon electrode (SPCE) with electrospun polysulfone (PS) nanofibers coated with dopamine. This imprinted sensor was used to measure NS1 in actual human serum samples and achieved satisfactory analytical performance with sensitivities ranging from 95 % to 97.14 % and standard deviations of less than five percent (5%). This sensor can detect NS1 concentrations as low as 0.3 ng/mL. In another study, an MIP-based electrochemical sensor was developed for the detection of COVID-19 [55]. This sensor uses a disposable thin-film metal electrode (Au-TFME) chip modified with MIP film encoded with a selective S1 component of the S protein (nCovS1) [55]. This sensor was evaluated using clinical samples and can detect nCovS1 from samples nasopharyngeal fluid within 15 min. The limit of detection (LOD) value ranges from 15 fM to 64 fM. Table 1 summarizes recent works on the electrochemical detection of COVID-19.
Table 1. Summary of electrochemical biosensor that are used for the detection of COVID-19 using protein, antibodies, nucleic acid and molecular imprinted polymer.
| Author | Instrument | Detection Method |
Analysis Time | Limit of Detection | Remarks |
|---|---|---|---|---|---|
| Alafeef et al. [40] |
Graphene-based electrochemical biosensor (integration of thiol-modified antisense oligonucleotide probes with AuNP caps) | Protein detection: nucleocapsid (N gene) of COVID-19 |
Less than 5 min | 6.9 copies/μL |
At present, the device is expected to be integrated with a portable mobile platform that can be used for instantaneous diagnosis of positive COVID-19 cases. |
| Vadlamani et al. [41] |
Cobalt-functionalized TiO2 nanotube (Co-TNT)-based electrochemical sensor | Protein detection: spike protein receptor-binding domain (RBD) |
~30 s | ~0.7 nM levels | This assay has a tendency to be applied for the diagnosis of other recognized respiratory viruses or in conjunction with a suitable metallic element to ensure TNT function. |
| Chaibun et al. [42] | Multiplex rolling circle amplification (RCA) | Protein detection: nucleocapsid and spike glycoprotein |
No data available | 1 copy/μL for both N and S genes | The RNA extraction procedure for this assay is currently being optimized, in addition to integration of a smartphone-based biosensor device. Researchers are also working on minimizing the turnaround time for the assay and simplifying the test method. |
| Yakoh et al. [43] |
Label-free, paper-based electrochemical platform | Antibody detection of IgM and IgG | 30 min | 1 ng/mL | Researchers are working on the direct detection of COVID-19 spike protein of with this assay. |
| Fan et al. [56] | Entropy-driven amplified electrochemiluminescence (ECL) biosensor | Nucleic acid detection includes the RdRp gene of COVID-19 | 30 min | As low as 2.67 fM | Not available |
| Ayankojo et al. [49] | MIP-based electrochemical sensor | S1 component of the S protein (nCovS1) | Not available | 4.8 pg/mL | This sensor demonstrates high selectivity to SARS-CoV-2 spike protein compared to other proteins. The sensor is compatible with portable potentiostats and suitable for point-of-care diagnosis. |
References
- Valizadeh, A.; Sohrabi, N.; Badrzadeh, F. Electrochemical detection of HIV-1 by nanomaterials. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1467–1477.
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128.
- Stat-i 400 Bipotentiostat/Galvanostat/Impedance Analyzer (EIS)|Metrohm. Available online: https://www.metrohm.com/en_my/products/s/tat-/stat-i-400.html (accessed on 8 July 2022).
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033.
- Antibody Types: IgM, IgA, IgD, IgG, IgE and Camelid Antibodies. Available online: https://www.news-medical.net/life-sciences/Types-of-Antibodies.aspx (accessed on 17 June 2022).
- Screen-Printed Gold Electrode (Aux.:Au; Ref.:Ag)/Ink AT/Work in Solution. Available online: https://www.metrohm.com/id_id/products/c/220a/c220at.html (accessed on 17 June 2022).
- Sekine, S.; Ido, Y.; Miyake, T.; Nagamine, K.; Nishizawa, M. Conducting Polymer Electrodes Printed on Hydrogel. J. Am. Chem. Soc. 2010, 132, 13174–13175.
- Chapman, C.A.R.; Cuttaz, E.A.; Tahirbegi, B.; Novikov, A.; Petkos, K.; Koutsoftidis, S.; Drakakis, E.M.; Goding, J.A.; Green, R.A. Flexible Networks of Patterned Conducting Polymer Nanowires for Fully Polymeric Bioelectronics. Adv. NanoBiomed. Res. 2022, 2, 2100102.
- Strnad, G.; German-Sallo, Z.; Jakab-Farkas, L.; Petrovan, C.; Portan, D. Influence of electrical parameters on morphology of nanostructured TiO2layers developed by electrochemical anodization. MATEC Web Conf. 2017, 112, 4021.
- Zhang, H.; Miller, B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: State of the art. Biosens. Bioelectron. 2019, 141, 111476.
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heeger, A.J. A Reagentless Signal-On Architecture for Electronic, Aptamer-Based Sensors via Target-Induced Strand Displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991.
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206.
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763.
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214.
- Batchelor-Mcauley, C.; Kätelhçn, E.; Barnes, E.O.; Compton, R.G.; Aborda, E.; Molina, A. Recent Advances in Voltammetry. ChemistryOpen 2015, 4, 224–260.
- Nossol, E.; Muñoz, R.A.A.; Richter, E.M.; de Souza Borges, P.H.; Silva, S.C.; Rocha, D.P. Sensing Materials: Graphene. Ref. Modul. Biomed. Sci. 2021.
- Harvey, D. Effect of Scan Rate in Cyclic Voltammetry. Available online: https://community.asdlib.org/imageandvideoexchangeforum/2013/07/31/effect-of-scan-rate-in-cyclic-voltammetry/ (accessed on 7 January 2022).
- Orazem, M.E.; Tribollet, B. Electrochemical Impedance Spectroscopy. New Jersey 2008, 383–389.
- Schroll, C.A.; Cohen, S.M. Electrochemical Impedance Spectroscopy. In Experiment-EIS-Teacher-Edition; Gamry Instruments Inc.: Warminster, PA, USA, 2016; ISBN 0-9983378-1-1.
- Sebastian, B.; Cg, T. Impedance, Electrical and Dielectric behaviour of Tin Oxide Nanoparticle doped with Graphite, Graphene Oxide and Reduced Graphene Oxide. Int. J. Electrochem. Sci. 2021, 16, 210810.
- Suni, I.I. Impedance methods for electrochemical sensors using nanomaterials. TrAC Trends Anal. Chem. 2008, 27, 604–611.
- Kaiser, G. 10.2: Size and Shapes of Viruses—Biology LibreTexts. Available online: https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Kaiser)/Unit_4%3A_Eukaryotic_Microorganisms_and_Viruses/10%3A_Viruses/10.02%3A_Size_and_Shapes_of_Viruses (accessed on 9 June 2022).
- Parker, N.; Schneegurt, M.; Tu, A.-H.T.; Forster, B.M.; Lister, P.; Allen, S.; Auman, A.; Brelles-Mariño, G.; Feldman, M.A.; Flowers, P.; et al. Microbiology; OpenStax: Houston, TX, USA, 2016; ISBN 1-947172-23-9.
- Navakul, K.; Warakulwit, C.; Yenchitsomanus, P.-T.; Panya, A.; Lieberzeit, P.A.; Sangma, C. A novel method for dengue virus detection and antibody screening using a graphene-polymer based electrochemical biosensor. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 549–557.
- Arshak, K.; Velusamy, V.; Korostynska, O.; Oliwa-Stasiak, K.; Adley, C. Conducting Polymers and Their Applications to Biosensors: Emphasizing on Foodborne Pathogen Detection. IEEE Sens. J. 2009, 9, 1942–1951.
- Guimard, N.K.; Gomez, N.; Schmidt, C.E. Conducting polymers in biomedical engineering. Prog. Polym. Sci. 2007, 32, 876–921.
- Wei, D.; Bailey, M.J.A.; Andrew, P.; Ryhänen, T. Electrochemical biosensors at the nanoscale. Lab Chip 2009, 9, 2123–2131.
- Singh, K.V.; Whited, A.M.; Ragineni, Y.; Barrett, T.W.; King, J.; Solanki, R. 3D nanogap interdigitated electrode array biosensors. Anal. Bioanal. Chem. 2010, 397, 1493–1502.
- Patolsky, F.; Lieber, C.M. Nanowire nanosensors. Mater. Today 2005, 8, 20–28.
- El Sharif, H.F.; Dennison, S.R.; Tully, M.; Crossley, S.; Mwangi, W.; Bailey, D.; Graham, S.P.; Reddy, S.M. Evaluation of electropolymerized molecularly imprinted polymers (E-MIPs) on disposable electrodes for detection of SARS-CoV-2 in saliva. Anal. Chim. Acta 2022, 1206, 339777.
- Jo, J.; Yoon, J.; Lee, T.; Cho, H.-Y.; Lee, J.-Y.; Choi, J.-W. H2O2 biosensor consisted of hemoglobin-DNA conjugate on nanoporous gold thin film electrode with electrochemical signal enhancement. Nano Converg. 2019, 6, 1.
- Dutta, N.; Lillehoj, P.B.; Estrela, P.; Dutta, G. Electrochemical Biosensors for Cytokine Profiling: Recent Advancements and Possibilities in the Near Future. Biosensors 2021, 11, 94.
- Songa, E.A.; Somerset, V.S.; Waryo, T.; Baker, P.G.L.; Iwuoha, E.I. Amperometric nanobiosensor for quantitative determination of glyphosate and glufosinate residues in corn samples. Pure Appl. Chem. 2009, 81, 123–139.
- Hong, P.; Li, W.; Li, J. Applications of Aptasensors in Clinical Diagnostics. Sensors 2012, 12, 1181–1193.
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557.
- Patris, S.; Vandeput, M.; Kauffmann, J.-M. Antibodies as target for affinity biosensors. TrAC Trends Anal. Chem. 2016, 79, 239–246.
- Cooper, M.A. Label-Free Biosensors: Techniques and Applications; Cambridge University Press: Cambridge, MA, USA, 2009; pp. 1–28.
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2015, 36, 465–481.
- Kumar, N.; Shetti, N.P.; Jagannath, S.; Aminabhavi, T.M. Electrochemical sensors for the detection of SARS-CoV-2 virus. Chem. Eng. J. 2022, 430, 132966.
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045.
- Vadlamani, B.S.; Uppal, T.; Verma, S.C.; Misra, M. Functionalized TiO2 Nanotube-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. Sensors 2020, 20, 5871.
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802.
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021, 176, 112912.
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, Solutions and Prospects. Acta Nat. 2013, 5, 34–43.
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403.
- Parashar, N.C.; Poddar, J.; Chakrabarti, S.; Parashar, G. Repurposing of SARS-CoV nucleocapsid protein specific nuclease resistant RNA aptamer for therapeutics against SARS-CoV-2. Infect. Genet. Evol. 2020, 85, 104497.
- Dalirirad, S.; Steckl, A.J. Lateral flow assay using aptamer-based sensing for on-site detection of dopamine in urine. Anal. Biochem. 2020, 596, 113637.
- Lum, J.; Wang, R.; Hargis, B.; Tung, S.; Bottje, W.; Lu, H.; Li, Y. An Impedance Aptasensor with Microfluidic Chips for Specific Detection of H5N1 Avian Influenza Virus. Sensors 2015, 15, 18565–18578.
- Ghanbari, K.; Roushani, M.; Azadbakht, A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017, 534, 64–69.
- Piletska, E.V.; Guerreiro, A.R.; Whitcombe, M.J.; Piletsky, S.A. Influence of the Polymerization Conditions on the Performance of Molecularly Imprinted Polymers. Macromolecules 2009, 42, 4921–4928.
- Scorrano, S.; Mergola, L.; Del Sole, R.; Vasapollo, G. Synthesis of Molecularly Imprinted Polymers for Amino Acid Derivates by Using Different Functional Monomers. Int. J. Mol. Sci. 2011, 12, 1735–1743.
- Chatterjee, T.N.; Bandyopadhyay, R. A Molecularly Imprinted Polymer-Based Technology for Rapid Testing of COVID-19. Trans. Indian Natl. Acad. Eng. 2020, 5, 225–228.
- Belbruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119.
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-Based Impedimetric Sensor for Detecting Dengue Fever Biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384–1394.
- Ayankojo, A.G.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sens. Actuators B Chem. 2022, 353, 131160.
- Fan, Z.; Yao, B.; Ding, Y.; Zhao, J.; Xie, M.; Zhang, K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021, 178, 113015.
More
