Repetitive sequences represent about half of the human genome. They are actively transcribed and play a role during development and in epigenetic regulation. The altered activity of repetitive sequences can lead to genomic instability and they can contribute to the establishment or the progression of degenerative diseases and cancer transformation. Increased levels of heterochromatic repetitive satellite-coded RNAs in mammary glands induce breast tumor formation in mice, altering the BRCA1-associated protein networks that are required for the proper stabilization of DNA replication forks that in turn lead to genomic instability. In humans, patients with breast cancer that express high levels of RNA derived from alpha satellite have an increased risk of developing multiple cancers.
- breast cancer
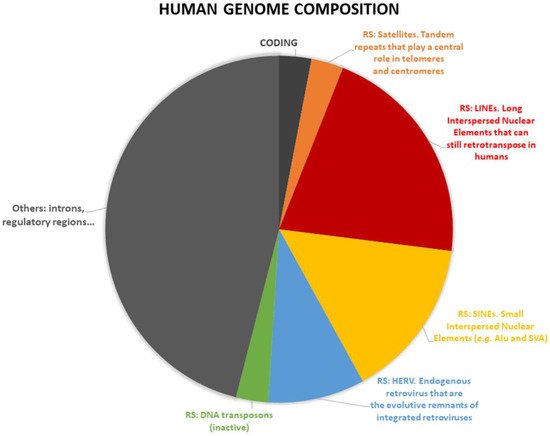
- repetitive sequences
- HERV
- endogenous retrovirus
- satellite repeats
- centromeres
- telomeres
- SVA
- LINE1
- transposons
1. Breast Cancer Classification
2. Non-Coding RNA in Mammals
3. Repetitive DNA Sequence Classification
4. Repetitive DNA Sequence and Cancer
References
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and Current Knowledge of Breast Cancer. Biol. Res. 2017, 50, 33.
- Tan, P.H.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 World Health Organization Classification of Tumours of the Breast. Histopathology 2020, 77, 181–185.
- Di Napoli, A.; Jain, P.; Duranti, E.; Margolskee, E.; Arancio, W.; Facchetti, F.; Alobeid, B.; Santanelli di Pompeo, F.; Mansukhani, M.; Bhagat, G. Targeted next Generation Sequencing of Breast Implant-Associated Anaplastic Large Cell Lymphoma Reveals Mutations in JAK/STAT Signalling Pathway Genes, TP53 and DNMT3A. Br. J. Haematol. 2018, 180, 741–744.
- Xu, S.; Kong, D.; Chen, Q.; Ping, Y.; Pang, D. Oncogenic Long Noncoding RNA Landscape in Breast Cancer. Mol. Cancer 2017, 16, 129.
- Pang, B.; Wang, Q.; Ning, S.; Wu, J.; Zhang, X.; Chen, Y.; Xu, S. Landscape of Tumor Suppressor Long Noncoding RNAs in Breast Cancer. J. Exp. Clin. Cancer Res. 2019, 38, 79.
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J. Strategies for Subtypes-Dealing with the Diversity of Breast Cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747.
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J.; Albain, K.S.; André, F.; Bergh, J.; et al. Personalizing the Treatment of Women with Early Breast Cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223.
- Hennigs, A.; Riedel, F.; Gondos, A.; Sinn, P.; Schirmacher, P.; Marmé, F.; Jäger, D.; Kauczor, H.U.; Stieber, A.; Lindel, K.; et al. Prognosis of Breast Cancer Molecular Subtypes in Routine Clinical Care: A Large Prospective Cohort Study. BMC Cancer 2016, 16, 734.
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical Implications of the Intrinsic Molecular Subtypes of Breast Cancer. Breast 2015, 24, S26–S35.
- Clark, M.B.; Amaral, P.P.; Schlesinger, F.J.; Dinger, M.E.; Taft, R.J.; Rinn, J.L.; Ponting, C.P.; Stadler, P.F.; Morris, K.V.; Morillon, A.; et al. The Reality of Pervasive Transcription. PLoS Biol. 2011, 9, e1000625.
- Arancio, W.; Coronnello, C. Repetitive Sequences in Aging. Aging 2021, 13, 10816–10817.
- Hirschberger, S.; Hinske, L.C.; Kreth, S. MiRNAs: Dynamic Regulators of Immune Cell Functions in Inflammation and Cancer. Cancer Lett. 2018, 431, 11–21.
- Li, M.; Cui, X.; Guan, H. MicroRNAs: Pivotal Regulators in Acute Myeloid Leukemia. Ann. Hematol. 2020, 99, 399–412.
- Liu, J.; Ke, F.; Chen, T.; Zhou, Q.; Weng, L.; Tan, J.; Shen, W.; Li, L.; Zhou, J.; Xu, C.; et al. MicroRNAs That Regulate PTEN as Potential Biomarkers in Colorectal Cancer: A Systematic Review. J. Cancer Res. Clin. Oncol. 2020, 146, 809–820.
- Arancio, W.; Calogero Amato, M.; Magliozzo, M.; Pizzolanti, G.; Vesco, R.; Giordano, C. Serum MiRNAs in Women Affected by Hyperandrogenic Polycystic Ovary Syndrome: The Potential Role of MiR-155 as a Biomarker for Monitoring the Estroprogestinic Treatment. Gynecol. Endocrinol. 2018, 34, 704–708.
- Hashemi, A.; Gorji-bahri, G. MicroRNA: Promising Roles in Cancer Therapy. Curr. Pharm. Biotechnol. 2020, 21, 1186–1203.
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived LncRNAs and Their MiRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 2020, 8, 85.
- Arancio, W.; Carina, V.; Pizzolanti, G.; Tomasello, L.; Pitrone, M.; Baiamonte, C.; Amato, M.C.; Giordano, C. Anaplastic Thyroid Carcinoma: A CeRNA Analysis Pointed to a Crosstalk between SOX2, TP53, and MicroRNA Biogenesis. Int. J. Endocrinol. 2015, 2015, 439370.
- Poliseno, L.; Pandolfi, P.P. PTEN CeRNA Networks in Human Cancer. Methods 2015, 77–78, 41–50.
- Arancio, W.; Genovese, S.I.; Bongiovanni, L.; Tripodo, C. A CeRNA Approach May Unveil Unexpected Contributors to Deletion Syndromes, the Model of 5q-Syndrome. Oncoscience 2015, 2, 872–879.
- Arancio, W.; Giordano, C.; Pizzolanti, G. A CeRNA Analysis on LMNA Gene Focusing on the Hutchinson-Gilford Progeria Syndrome. J. Clin. Bioinform. 2013, 3, 2.
- Arancio, W. A Bioinformatics Analysis of Lamin-A Regulatory Network: A Perspective on Epigenetic Involvement in Hutchinson-Gilford Progeria Syndrome. Rejuvenation Res. 2012, 15, 123–127.
- Arancio, W. CeRNA Analysis of SARS-CoV-2. Arch. Virol. 2021, 166, 271–274.
- Bertolazzi, G.; Cipollina, C.; Benos, P.V.; Tumminello, M.; Coronnello, C. MiR-1207-5p Can Contribute to Dysregulation of Inflammatory Response in COVID-19 via Targeting SARS-CoV-2 RNA. Front. Cell. Infect. Microbiol. 2020, 10, 586592.
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The Regulated Retrotransposon Transcriptome of Mammalian Cells. Nat. Genet. 2009, 41, 563–571.
- Clayton, E.A.; Wang, L.; Rishishwar, L.; Wang, J.; McDonald, J.F.; Jordan, I.K. Patterns of Transposable Element Expression and Insertion in Cancer. Front. Mol. Biosci. 2016, 3, 76.
- Sciamanna, I.; De Luca, C.; Spadafora, C. The Reverse Transcriptase Encoded by LINE-1 Retrotransposons in the Genesis, Progression, and Therapy of Cancer. Front. Chem. 2016, 4, 6.
- Ohms, S.; Lee, S.H.; Rangasamy, D. LINE-1 Retrotransposons and Let-7 MiRNA: Partners in the Pathogenesis of Cancer? Front. Genet. 2014, 5, 338.
- Tubio, J.M.C.; Li, Y.; Ju, Y.S.; Martincorena, I.; Cooke, S.L.; Tojo, M.; Gundem, G.; Pipinikas, C.P.; Zamora, J.; Raine, K.; et al. Extensive Transduction of Nonrepetitive DNA Mediated by L1 Retrotransposition in Cancer Genomes. Science 2014, 345, 1251343.
- Di Ruocco, F.; Basso, V.; Rivoire, M.; Mehlen, P.; Ambati, J.; De Falco, S.; Tarallo, V. Alu RNA Accumulation Induces Epithelial-to-Mesenchymal Transition by Modulating MiR-566 and Is Associated with Cancer Progression. Oncogene 2018, 37, 627–637.
- Padeken, J.; Zeller, P.; Gasser, S.M. Repeat DNA in Genome Organization and Stability. Curr. Opin. Genet. Dev. 2015, 31, 12–19.
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and Next-Generation Sequencing: Computational Challenges and Solutions. Nat. Rev. Genet. 2012, 13, 36–46.
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 Retrotransposition in Human Neural Progenitor Cells. Nature 2009, 460, 1127–1131.
- Arancio, W. Progerin Expression Induces a Significant Downregulation of Transcription from Human Repetitive Sequences in IPSC-Derived Dopaminergic Neurons. GeroScience 2019, 41, 39–49.
- Zhu, Q.; Hoong, N.; Aslanian, A.; Hara, T.; Benner, C.; Heinz, S.; Miga, K.H.; Ke, E.; Verma, S.; Soroczynski, J.; et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cell 2018, 70, 842–853.e7.
- Kakizawa, N.; Suzuki, K.; Abe, I.; Endo, Y.; Tamaki, S.; Ishikawa, H.; Watanabe, F.; Ichida, K.; Saito, M.; Futsuhara, K.; et al. High Relative Levels of Satellite Alpha Transcripts Predict Increased Risk of Bilateral Breast Cancer and Multiple Primary Cancer in Patients with Breast Cancer and Lacking BRCA-Related Clinical Features. Oncol. Rep. 2019, 42, 857–865.
- Patnala, R.; Lee, S.H.; Dahlstrom, J.E.; Ohms, S.; Chen, L.; Dheen, S.T.; Rangasamy, D. Inhibition of LINE-1 Retrotransposon-Encoded Reverse Transcriptase Modulates the Expression of Cell Differentiation Genes in Breast Cancer Cells. Breast Cancer Res. Treat. 2014, 143, 239–253.
- Park, S.Y.; Seo, A.N.; Jung, H.Y.; Gwak, J.M.; Jung, N.; Cho, N.Y.; Kang, G.H. Alu and LINE-1 Hypomethylation Is Associated with HER2 Enriched Subtype of Breast Cancer. PLoS ONE 2014, 9, e100429.
- Van Hoesel, A.Q.; Van De Velde, C.J.H.; Kuppen, P.J.K.; Liefers, G.J.; Putter, H.; Sato, Y.; Elashoff, D.A.; Turner, R.R.; Shamonki, J.M.; De Kruijf, E.M.; et al. Hypomethylation of LINE-1 in Primary Tumor Has Poor Prognosis in Young Breast Cancer Patients: A Retrospective Cohort Study. Breast Cancer Res. Treat. 2012, 134, 1103–1114.
- Harris, C.R.; Normart, R.; Yang, Q.; Stevenson, E.; Haffty, B.G.; Ganesan, S.; Cordon-Cardo, C.; Levine, A.J.; Tang, L.H. Association of Nuclear Localization of a Long Interspersed Nuclear Element-1 Protein in Breast Tumors with Poor Prognostic Outcomes. Genes and Cancer 2010, 1, 115–124.
- Gualtieri, A.; Andreola, F.; Sciamanna, I.; Sinibaldi-Vallebona, P.; Serafino, A.; Spadafora, C. Increased Expression and Copy Number Amplification of LINE-1 and SINE B1 Retrotransposable Elements in Murine Mammary Carcinoma Progression. Oncotarget 2013, 4, 1882–1893.
- Chen, L.; Dahlstrom, J.E.; Chandra, A.; Board, P.; Rangasamy, D. Prognostic Value of LINE-1 Retrotransposon Expression and Its Subcellular Localization in Breast Cancer. Breast Cancer Res. Treat. 2012, 136, 129–142.
- Miret, N.; Zappia, C.D.; Altamirano, G.; Pontillo, C.; Zárate, L.; Gómez, A.; Lasagna, M.; Cocca, C.; Kass, L.; Monczor, F.; et al. AhR Ligands Reactivate LINE-1 Retrotransposon in Triple-Negative Breast Cancer Cells MDA-MB-231 and Non-Tumorigenic Mammary Epithelial Cells NMuMG. Biochem. Pharmacol. 2020, 175, 113904.
- Miglio, U.; Berrino, E.; Panero, M.; Ferrero, G.; Coscujuela Tarrero, L.; Miano, V.; Dell’Aglio, C.; Sarotto, I.; Annaratone, L.; Marchiò, C.; et al. The Expression of LINE1-MET Chimeric Transcript Identifies a Subgroup of Aggressive Breast Cancers. Int. J. Cancer 2018, 143, 2838–2848.
- Bratthauer, G.L.; Cardiff, R.D.; Fanning, T.G. Expression of LINE-1 Retrotransposons in Human Breast Cancer. Cancer 1994, 73, 2333–2336.
- Wang, Y.; Bernhardy, A.J.; Nacson, J.; Krais, J.J.; Tan, Y.F.; Nicolas, E.; Radke, M.R.; Handorf, E.; Llop-Guevara, A.; Balmaña, J.; et al. BRCA1 Intronic Alu Elements Drive Gene Rearrangements and PARP Inhibitor Resistance. Nat. Commun. 2019, 10, 5661.
- Staaf, J.; Glodzik, D.; Bosch, A.; Vallon-Christersson, J.; Reuterswärd, C.; Häkkinen, J.; Degasperi, A.; Amarante, T.D.; Saal, L.H.; Hegardt, C.; et al. Whole-Genome Sequencing of Triple-Negative Breast Cancers in a Population-Based Clinical Study. Nat. Med. 2019, 25, 1526–1533.
- Felicio, P.S.; Alemar, B.; Coelho, A.S.; Berardinelli, G.N.; Melendez, M.E.; Lengert, A.V.H.; Miche lli, R.D.; Reis, R.M.; Fernandes, G.C.; Ewald, I.P.; et al. Screening and Characterization of BRCA2 c.156_157insAlu in Brazil: Results from 1380 Individuals from the South and Southeast. Cancer Genet. 2018, 228–229, 93–97.
- Rizza, R.; Hackmann, K.; Paris, I.; Minucci, A.; De Leo, R.; Schrock, E.; Urbani, A.; Capoluongo, E.; Gelli, G.; Concolino, P. Novel BRCA1 Large Genomic Rearrangements in Italian Breast/Ovarian Cancer Patients. Mol. Diagnosis Ther. 2019, 23, 121–126.
- Gallegos-Arreola, M.P.; Figuera, L.E.; Flores-Ramos, L.G.; Puebla-Pérez, A.M.; Zúñiga-González, G.M. Association of the Alu Insertion Polymorphism in the Progesterone Receptor Gene with Breast Cancer in a Mexican Population. Arch. Med. Sci. 2015, 11, 551–560.
- Machado, P.M.; Brandão, R.D.; Cavaco, B.M.; Eugénio, J.; Bento, S.; Nave, M.; Rodrigues, P.; Fernandes, A.; Vaz, F. Screening for a BRCA2 Rearrangement in High-Risk Breast/Ovarian Cancer Families: Evidence for a Founder Effect and Analysis of the Associated Phenotypes. J. Clin. Oncol. 2007, 25, 2027–2034.
- Wu, Y.; Zhao, W.; Liu, Y.; Tan, X.; Li, X.; Zou, Q.; Xiao, Z.; Xu, H.; Wang, Y.; Yang, X. Function of HNRNPC in Breast Cancer Cells by Controlling the DsRNA-induced Interferon Response. EMBO J. 2018, 37, e99017.
- Grabski, D.F.; Hu, Y.; Sharma, M.; Rasmussen, S.K. Close to the Bedside: A Systematic Review of Endogenous Retroviruses and Their Impact in Oncology. J. Surg. Res. 2019, 240, 145–155.
- Zhao, J.; Rycaj, K.; Geng, S.; Li, M.; Plummer, J.B.; Yin, B.; Liu, H.; Xu, X.; Zhang, Y.; Yan, Y.; et al. Expression of Human Endogenous Retrovirus Type K Envelope Protein Is a Novel Candidate Prognostic Marker for Human Breast Cancer. Genes Cancer 2011, 2, 914–922.
- Wang-Johanning, F.; Radvanyi, L.; Rycaj, K.; Plummer, J.B.; Yan, P.; Sastry, K.J.; Piyathilake, C.J.; Hunt, K.K.; Johanning, G.L. Human Endogenous Retrovirus K Triggers an Antigen-Specific Immune Response in Breast Cancer Patients. Cancer Res. 2008, 68, 5869–5877.
- Golan, M.; Hizi, A.; Resau, J.H.; Yaal-Hahoshen, N.; Reichman, H.; Keydar, I.; Tsarfaty, I. Human Endogenous Retrovirus (HERV-K) Reverse Transcriptase as a Breast Cancer Prognostic Marker. Neoplasia 2008, 10, 521–533.
- Wang-Johanning, F.; Frost, A.R.; Jian, B.; Epp, L.; Lu, D.W.; Johanning, G.L. Quantitation of HERV-K Env Gene Expression and Splicing in Human Breast Cancer. Oncogene 2003, 22, 1528–1535.
- Wang-Johanning, F.; Frost, A.R.; Johanning, G.L.; Khazaeli, M.B.; LoBuglio, A.F.; Shaw, D.R.; Strong, T.V. Expression of Human Endogenous Retrovirus K Envelope Transcripts in Human Breast Cancer. Clin. Cancer Res. 2001, 7, 1553–1560.
- Matteucci, C.; Balestrieri, E.; Argaw-Denboba, A.; Sinibaldi-Vallebona, P. Human Endogenous Retroviruses Role in Cancer Cell Stemness. Semin. Cancer Biol. 2018, 53, 17–30.
- Salmons, B.; Lawson, J.S.; Günzburg, W.H. Recent Developments Linking Retroviruses to Human Breast Cancer: Infectious Agent, Enemy within or Both? J. Gen. Virol. 2014, 95, 2589–2593.
- Nguyen, T.D.; Davis, J.; Eugenio, R.A.; Liu, Y. Female Sex Hormones Activate Human Endogenous Retrovirus Type K Through the OCT4 Transcription Factor in T47D Breast Cancer Cells. AIDS Res. Hum. Retrovir. 2019, 35, 348–356.
- Johanning, G.L.; Malouf, G.G.; Zheng, X.; Esteva, F.J.; Weinstein, J.N.; Wang-Johanning, F.; Su, X. Expression of Human Endogenous Retrovirus-K Is Strongly Associated with the Basal-like Breast Cancer Phenotype. Sci. Rep. 2017, 7, 41960.
- Jin, X.; Xu, X.E.; Jiang, Y.Z.; Liu, Y.R.; Sun, W.; Guo, Y.J.; Ren, Y.X.; Zuo, W.J.; Hu, X.; Huang, S.L.; et al. The Endogenous Retrovirus-Derived Long Noncoding RNA TROJAN Promotes Triple-Negative Breast Cancer Progression via ZMYND8 Degradation. Sci. Adv. 2019, 5, eaat9820.
- Lemaître, C.; Tsang, J.; Bireau, C.; Heidmann, T.; Dewannieux, M. A Human Endogenous Retrovirus-Derived Gene That Can Contribute to Oncogenesis by Activating the ERK Pathway and Inducing Migration and Invasion. PLoS Pathog. 2017, 13, e1006451.
- Zhou, F.; Li, M.; Wei, Y.; Lin, K.; Lu, Y.; Shen, J.; Johanning, G.L.; Wang-Johanning, F. Activation of HERV-K Env Protein Is Essential for Tumorigenesis and Metastasis of Breast Cancer Cells. Oncotarget 2016, 7, 84093–84117.
- Wang-Johanning, F.; Li, M.; Esteva, F.J.; Hess, K.R.; Yin, B.; Rycaj, K.; Plummer, J.B.; Garza, J.G.; Ambs, S.; Johanning, G.L. Human Endogenous Retrovirus Type K Antibodies and MRNA as Serum Biomarkers of Early-Stage Breast Cancer. Int. J. Cancer 2014, 134, 587–595.
- Jiang, J.C.; Upton, K.R. Human Transposons Are an Abundant Supply of Transcription Factor Binding Sites and Promoter Activities in Breast Cancer Cell Lines. Mob. DNA 2019, 10, 16.
- Presneau, N.; Laplace-Marieze, V.; Sylvain, V.; Lortholary, A.; Hardouin, A.; Bernard-Gallon, D.; Bignon, Y.J. New Mechanism of BRCA-1 Mutation by Deletion/Insertion at the Same Nucleotide Position in Three Unrelated French Breast/Ovarian Cancer Families. Hum. Genet. 1998, 103, 334–339.
