The ecosystem of the human gastrointestinal tract, named gut microbiota, represents the most thoroughly mapped ecosystem. Perturbations on bacterial populations cause dysbiosis, a condition correlated to a wide range of autoimmune, neurological, metabolic, cardiovascular, and respiratory diseases. The lungs have their flora, which are directly related to the gut flora via bidirectional communication allowing the transport of microbial metabolites and toxins produced by intestinal bacteria through the circulation and lymphatic system. This mutual microbial cross-talk communication called the gut–lung axis modulates the immune and inflammatory response to infections. COVID-19 causes dysbiosis, altered intestinal permeability, and bacterial translocation. Dysbiosis, through the gut–lung axis, promotes hyper-inflammation, exacerbates lung damage, and worsens clinical outcomes.
1. Introduction
The gastrointestinal tract is colonized by a wide range of dynamic microbial populations, consisting mainly of anaerobic bacteria, known as gut microbiota. Direct microbial–host interaction affects many physiological functions such as digestion, metabolism, intestinal barrier integrity (epithelial barrier), organ function, and immune homeostasis
[1]. Bacteria usually live in a symbiotic relationship with the host, while perturbations in their composition and function cause dysbiosis associated with gastrointestinal, neurological, metabolic, cardiovascular, and respiratory disorders
[2]. Lungs, for years, have been considered to be sterile because microbiological culture techniques have shown negative results. Nowadays, technological advances confirm that healthy lungs host a dynamic and diverse bacterial ecosystem
[3]. The predominant genera that colonize healthy lungs are Bacteroidetes and Firmicutes, followed by Proteobacteria and Actinobacteria
[4]. Dysbiosis can weaken nonspecific lung immunity, promote the host’s susceptibility to infections, and may worsen chronic lung disease. In addition, changes in lung flora affect cytokine production by triggering the transition from anti-inflammatory to proinflammatory cytokines, thus promoting injury to the pulmonary epithelium resulting in fibrosis
[4]. The interaction between lung and gut microbiota involves a bidirectional communication with metabolites and toxins produced by intestinal bacteria arriving at the lungs through the circulation and lymphatic system, known as the “gut-lung axis”
[5]. Microbiota-immune interactions regulate the inflammatory response by activating this axis through bacterial populations and their by-products which leak the mucosal barrier (translocation)
[2]. The bacterial translocation, leukocyte transport, and inflammatory cytokine secretion through the gut–lung axis are phenomena also found in SARS-CoV-2 infection, worsening the clinical outcome of patients
[6]. Although COVID-19 is primarily a viral respiratory disease caused by SARS-CoV-2, more and more evidence suggests that the gastrointestinal tract is involved in COVID-19
[7]. SARS-CoV-2 infects intestinal epithelial cells (IECs) too, due to its transport through the lymphatic system, resulting in disruption of intestinal permeability and mucosal integrity
[8]. Thus, there appears to be a vital cross-communication between the microbial communities in the mucous membranes of the lungs and the intestine
[4]. Probiotics have well-documented antiviral activities against common respiratory viruses such as respiratory syncytial virus, influenza, and rhinovirus. Probiotics regulate cytokine secretion, thus affecting both nonspecific and specific immune response. Bacteriocins, produced by lactic acid bacteria (LAB), are proteinaceous substances with antimicrobial properties that can inhibit the adhesion and invasion of other agents in the intestinal epithelium
[9], as shown in
Figure 1.
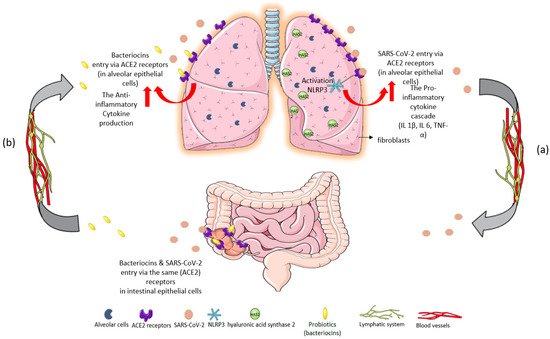
Figure 1. Immunomodulatory response of gut–lung axis cross-talk after: (
a) SARS-CoV-2 invasion and (
b) probiotic intake of
L. plantarum. (
a) SARS-CoV-2 binds to ACE2 receptors in the epithelial cells of the lungs. The massive cytokine release after viral entry leads to NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) inflammasome overactivation, exacerbating the inflammatory cascade
[9]. High levels of proinflammatory cytokines IL-1β, IL-6, and TNF-α are inducers of hyaluronic acid synthase 2 (HAS2) in the endothelium, alveolar epithelial cells, and fibroblasts. Hyperinflammation leads to hyaluronic acid production and accumulation in the alveoli of patients with severe COVID-19. The development of pulmonary fibrosis occurs due to inflammatory response and severe infection. The virus infects intestinal epithelial cells in the same way that it directly attacks alveolar epithelial cells. (
b) Probiotics could restrict SARS-CoV-2 invasion and proliferation through their metabolites, bacteriocins (antimicrobial peptides), because of their ability to block ACE2 receptors
[9]. Specifically,
L. plantarum bacteriocins compete with the virus for the same host sites
[10]. Thus, probiotics could offer a protective effect against acute respiratory distress syndrome (ARDS) in COVID-19 infection, reverse the “cytokine storm”, and reduce hyaluronic acid synthesis in the lungs
[11][12][11,12].
2. Preclinical Trials Supporting the Gut–Lung Axis Communication in COVID-19
Sencio et al. used Syrian hamsters for their exceptional ability to mimic the onset of pathological manifestations of COVID-19 disease. The infected animals had an abundance of opportunistic pathogenic bacteria, such as
Enterobacteriaceae and
Desulfovibrionaceae, a decrease in beneficial Short-Chain Fatty Acids (SCFAs)-producing
Ruminococcaceae and
Lachnospiraceae, and reduced concentration of SCFAs in the blood. Since SCFAs have a dual function, namely, a protective role in maintaining the integrity of intestinal epithelial barrier and regulating inflammatory response, the above findings were considered compatible with dysbiosis. However, SCFAs supplements failed to improve the clinical outcomes
[13].
Studying primates, Sokol et al. infected macaques with SARS-CoV-2 and observed alterations in the gut flora composition, which peaked on the 13th day postinfection. The relative abundance of the genus
Acinetobacter (Proteobacteria) and the family
Ruminococcaceae (Firmicutes) were associated with viral presence in the upper respiratory tract, and
Peptostreptococcaceae (especially of the Intestinibacter) was so in the rectum. These bacterial strains were associated with increased inflammatory markers in blood samples (C-reactive protein and proinflammatory cytokines). Additionally, a positive correlation was identified among several species of the genus
Streptococcus and chemokine production after viral infection. On the other hand, SARS-CoV-2 reduced SCFA levels in macaque feces, which affected the metabolism of tryptophan, a precursor of serotonin, and significantly altered the synthesis of various metabolites and bile acids. It remains unclear if the decrease in SCFAs concentrations may be related to lower production from gut bacteria or higher needs from the host cells. Researchers concluded that the reduced SCFA levels may be associated with proinflammatory conditions, increasing susceptibility to viral respiratory infections, including COVID-19, while the infected macaques developed mild symptoms of pneumonia, similar to those in people with COVID-19
[1].
Groves et al. evaluated the effect of a viral lung infection on gut flora composition by replicating infection with common respiratory pathogens such as respiratory syncytial virus (RSV) and influenza virus in mice. Animals were inoculated with a vaccine strain of live attenuated influenza virus (LAIV). Results after inoculation showed that RSV and influenza infection led to significant changes in the diversity of the intestinal flora, with an increase in Bacteroidetes phylum and a concomitant decrease in the abundance of Firmicutes. Although Bacteroidetes growth was consistently repeatable across multiple experiments, differences were observed in the bacterial family and taxonomy level. This fact suggests a change in the gut environment following a viral respiratory infection, which favors the growth of the general Bacteroidetes population, but not individual bacterial families. No change in gut flora composition was observed after vaccination with live attenuated influenza vaccine (LAIV), suggesting that the cause of gut microbiota change is related to the virulence of the viral strain. Viral respiratory infections also led to increased lipocalin-2 (a biomarker of gut inflammation) in stool, indicating low-grade gut inflammation and increased levels of Muc5ac mucin in both the airway mucosa and the colon. This study highlights the gut microbiota alterations after viral respiratory infections, whereas these changes were not observed during vaccination. Whether increased mucus levels and gut inflammation are the cause or effect of these changes is not yet clear
[14].
3. Clinical Trials Supporting the Gut–Lung Axis Communication in COVID-19
Although COVID-19 is primarily considered a respiratory infection, there is growing evidence supporting the GI involvement in the disease. Jin et al. in China analyzed 74 (11.4%) of 651 patients with COVID-19 who had at least one gastrointestinal (GI) tract symptom (nausea, vomiting, and diarrhea). Diarrhea was the most common symptom, accounting for 8.14% of the enrolled patients. Jin et al. reported, for the first time, novel characteristics of SARS-CoV-2 and its ability to infect the GI tract
[15].
In Hong Kong, fecal samples from 15 patients with COVID-19 were analyzed and microbiome data were compared with those from 6 subjects with community-acquired pneumonia and 15 healthy individuals (controls). Patients with COVID-19 had significant alterations in gut microbiota compared to controls, characterized by an abundance of opportunistic pathogens and a reduction of beneficial microbes during hospitalization. Gut dysbiosis persisted even after SARS-CoV-2 and the resolution of respiratory symptoms. The baseline abundance of
Coprobacillus,
Clostridium ramosum, and
Clostridium hathewayi correlated with COVID-19 severity. There was an inverse correlation between the higher populations of
Faecalibacterium prausnitzii (an anti-inflammatory bacterium) and disease severity. During the hospitalization,
Bacteroides dorei,
Bacteroides thetaiotaomicron,
Bacteroides massiliensis, and
Bacteroides ovatus, which reduce ACE2 expression in the murine gut, were correlated inversely with SARS-CoV-2 load in patients’ fecal samples
[16].
Tao Zuo et al. collected fecal samples from patients with COVID-19 during hospitalization until discharge. Subsequently, they compared fecal mycobiome compositions of 30 patients with those of 9 subjects with community-acquired pneumonia and 30 healthy individuals (controls) and assessed their profiles during hospitalization until viral clearance. Samples from patients with COVID-19 were abundant in opportunistic pathogens, such as
Candida albicans,
Candida auris, and
Aspergillus flavus, compared to controls.
A. flavus and
Aspergillus niger, two respiratory-associated fungal pathogens, were detected in fecal samples from a subgroup of patients with COVID-19, even after clearance of SARS-CoV-2 from nasopharyngeal samples and resolution of respiratory symptoms supporting the gut–lung axis hypothesis
[17]. In another pilot study, the same research group provided evidence from 7 of 15 patients with COVID-19 who had stool positivity for SARS-CoV-2 by viral RNA metagenomic sequencing even in the absence of GI manifestations. Patients had active and prolonged quiescent GI infection even after recovery from respiratory symptoms of SARS-CoV-2. The gut microbiota of these patients were characterized by enrichment of opportunistic pathogens
Collinsella aerofaciens,
Collinsella tanakaei,
Streptococcus infantis,
Morganella morganii, and loss of beneficial bacteria
[18].
Yeoh et al. collected and analyzed blood samples from 100 patients and stool samples from 27 of the total patients up to 30 days after clearance of SARS-CoV-2. Then, they measured inflammatory cytokines and blood biomarkers in blood plasma. Results showed increased IL-10, TNF-α, and biomarkers such as C reactive protein, lactate dehydrogenase, aspartate aminotransferase, and gamma-glutamyl transferase. Gut flora were significantly altered in patients with COVID-19 compared to healthy controls. Bacteria with immunomodulatory activity, such as
Faecalibacterium prausnitzii,
Eubacterium rectale, and Bifidobactria, were diminished and remained reduced 30 days after recovery. Alterations in gut microbiota were associated with disease severity
[19].
Li et al. examined the gut microbiota of 47 patients with COVID-19 and compared them with those of healthy controls (n = 19). Fecal samples showed four bacterial species,
Streptococcus thermophilus,
Bacteroides oleiciplenus,
Fusobacterium ulcerans, and
Prevotella bivia. Bacterial strains of
Bacteroides stercoris,
B. vulgatus,
B. massiliensis,
Bifidobacterium longum,
Streptococcus thermophilus,
Lachnospiraceae bacterium 5163FAA,
Prevotella bivia,
Erysipelotrichaceae bacterium Erysipelotrichaceae 6145, and
Erysipelotrichaceae 2244A were found in abundance in COVID-19 patients, while
Clostridium nexile,
Streptococcus salivarius,
Coprococcus catus,
Eubacterium hallii,
Enterobacter aerogenes, and
Adlercreutzia equolifaciens were reduced. The relative abundance of butyrate-producing
Roseburia inulinivorans was markedly reduced in COVID-19 patients, while
Paraprevotella sp. and
Streptococcus thermophilus were increased. This study showed that
Roseburia inulinivorans,
Bacteroides faecis,
Bifidobacterium bifidum,
Parabacteroides goldsteinii,
Lachnospiraceae bacterium 9143BFAA, and
Megasphaera sp. Had no correlation with disease severity, while
Paraprevotella sp.,
Streptococcus thermophilus,
Clostridium ramosum, and
Bifidobacterium animalis were positively correlated with the severity of COVID-19
[20].
In another study, Vestad et al. collected blood plasma from patients on hospital admission and three months after severe COVID-19 for the assessment of inflammatory markers and intestinal barrier dysfunction. During the follow-up, pulmonary function was assessed by measuring the diffusing capacity of the lungs for carbon monoxide (DL
CO). Rectal swabs were collected for gut microbiota analysis by sequencing the 16S rRNA gene. Results showed that gut flora decreased in COVID-19 patients with respiratory dysfunction (defined as the ability to DL
CO below the threshold three months after hospitalization). Specifically, decreased abundance of the family
Erysipelotrichaceae UCG-003 and increased
Flavonifractor and
Veillonella species were observed, with
Veillonella being possibly associated with fibrosis of the lung parenchyma. During hospitalization, elevated levels of lipopolysaccharide-binding protein (LBP) in patients’ blood plasma were directly correlated with respiratory failure, defined as pO
2/fiO
2 < 26.6 kPa. LBP levels remained elevated during and after three months of hospitalization and were associated with low-grade inflammation and respiratory dysfunction. Respiratory dysfunction after COVID-19 was associated with disturbances in gut flora, which enhance the gut–lung axis hypothesis
[7].