The outbreak of a novel coronavirus SARS-CoV-2 responsible for the COVID-19 pandemic has caused a worldwide public health emergency. Due to the constantly evolving nature of the coronaviruses, SARS-CoV-2-mediated alterations on post-transcriptional gene regulations across human tissues remain elusive. In this study, we analyzed publicly available genomic datasets to systematically dissect the crosstalk and dysregulation of the human post-transcriptional regulatory networks governed by RNA-binding proteins (RBPs) and micro-RNAs (miRs) due to SARS-CoV-2 infection. We uncovered that 13 out of 29 SARS-CoV-2-encoded proteins directly interacted with 51 human RBPs, of which the majority of them were abundantly expressed in gonadal tissues and immune cells. We further performed a functional analysis of differentially expressed genes in mock-treated versus SARS-CoV-2-infected lung cells that revealed enrichment for the immune response, cytokine-mediated signaling, and metabolism-associated genes. This study also characterized the alternative splicing events in SARS-CoV-2-infected cells compared to the control, demonstrating that skipped exons and mutually exclusive exons were the most abundant events that potentially contributed to differential outcomes in response to the viral infection. A motif enrichment analysis on the RNA genomic sequence of SARS-CoV-2 clearly revealed the enrichment for RBPs such as SRSFs, PCBPs, ELAVs, and HNRNPs, suggesting the sponging of RBPs by the SARS-CoV-2 genome.
- SARS-CoV-2
- COVID-19
- post-transcriptional regulation
- RNA Binding Proteins
- microRNAs
- RNA
- Network biology
1. Introduction
An outbreak of coronavirus disease (COVID-19) caused by the newly discovered severe acute respiratory syndrome coronavirus (SARS-CoV-2) started in December 2019 in the city of Wuhan, Hubei Province, China. As of September 16, 2020, COVID-19 has expanded globally, with more than 30 million confirmed cases with over 944,000 deaths worldwide, imposing an unprecedented threat to the public health (https://www.worldometers.info/coronavirus/). In the past two decades, coronavirus outbreaks have resulted in viral epidemics, including a severe acute respiratory syndrome (SARS-CoV) in 2002 with a fatality of 10% and the Middle East respiratory syndrome (MERS-CoV) in 2012 with fatality of 36% [1][2][3][4]. Both SARS-CoV and MERS-CoV were zoonotic viruses originating in bats and camels, respectively [5][6]. However, the recurring emergence of highly pathogenic SARS-CoV, MERS-CoV, and now SARS-CoV-2 have indicated the potential for cross-species transmission of these viruses, thus raising a serious public health concern [7][8]. SARS CoV-2 shares a sequence similarity of 80% and 50% with previously identified SARS-CoV and MERS-CoV, respectively [9][10][11][12]. Since its emergence, rapid efforts have illustrated the molecular features of SARS-CoV-2 that enable it to hijack the host cellular machinery and facilitates its genomic replication and assembly into new virions during the infection process [13][14][15][16].
Coronavirus carries the largest genome among all RNA viruses, ranging from 26 to 32 kilobases in length [12] This virus has a characteristic “crown”-like appearance under two-dimensional transmission electron microscopy. SARS-CoV-2 is an enveloped positive-sense, single-stranded ribonucleic acid (RNA) coronavirus that belongs to the genus beta-coronavirus. Upon entry in the cell, SARS-CoV-2 RNA is translated into nonstructural proteins (nsps) from two open reading frames (ORFs): ORF1a and ORF1b [17][18]. The ORF1a produces polypeptide 1a, which is cleaved further into 11 nsps, while ORF1b yields polypeptide 1ab, which is cleaved into 16 nsps [17][18]. Since SARS-CoV-2 utilizes human machinery to translate its RNA after entry into the cell, it could possibly impact several RNA-binding proteins from the host to bind the viral genome, resulting in altered post-transcriptional regulation. Next, the viral genome is used as the template for replication and transcription, mediated by nonstructural protein RNA-dependent RNA polymerase (RdRP) [18][19]. SARS-CoV-2 encodes four main structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N), which are conserved, and several other accessory proteins (3a, 6, 7a, 7b, 8, and 10), according to the current annotation (GenBank: NC_045512.2) [17][20]. The spike protein, which has evolved the most during the COVID-19 outbreak, enables the virus to bind to angiotensin-converting enzyme 2 (ACE2) on the host cell membrane, following which, it undergoes structural changes and, subsequently, allows the viral genome to make its way inside the host cell [21]. Infections caused by these viruses result in severe pneumonia, fever, and breathing difficulty [22].
A protein-protein interaction map between SARS-CoV-2 and the human proteins published recently has revealed several important targets for drug repurposing [23]. Given the evolving nature of coronaviruses that results in frequent genetic diversity in their genome, it is crucial to identify the regulators in humans that interact with the viral genome and their crosstalk that results in altered regulatory mechanisms in the host during the infection process. Therefore, it is imperative to investigate the interacting post-transcriptional regulators that asset these viral proteins in different tissues.
RNA-binding proteins (RBPs) are a class of proteins in humans that bind to single- or double-stranded RNA and facilitate the formation of ribonucleoprotein complexes [[24][25][26]. In addition to RBPs, micro-RNAs (miRs) that belong to a class of noncoding RNAs also interact with target RNAs to regulate the cognate RNA expression [27][28]. Both RBPs and miRs have been widely recognized in regulating the post-transcriptional gene regulatory network in humans [29][30][31]. Dysregulated RBPs and miRs have been shown to contribute significantly to the altered regulatory network in a plethora of diseases, such as cancer, genetic diseases, and viral infections [32][33][34][35][36][37][38]. Previous studies have shown that human RBPs, including the heterogeneous Nuclear Ribonucleoprotein family (hnRNPA1 and hnRNPAQ), polypyrimidine tract-binding protein (PTB), Serine/Arginine-Rich Splicing Factor 7 (SRSF7), and Transformer 2 Alpha Homolog (TRA2A), interact with coronavirus RNA [39][40][41][42][43][44]. Likewise, other reports have demonstrated the potential interaction between human miRNA and the viral genome, including a variety of coronaviruses [41][45][46]. However, the potential RBPs and miRs that interact with SARS-CoV-2 and their implications in viral pathogenesis has been poorly understood.
Currently, there are no proven antiviral therapeutics that are effective against the novel coronavirus. Although the analysis of therapeutic targets for SARS-CoV-2 has been conducted to identify potential drugs by computational methods [47], the targets have not been clinically approved for therapeutic applications. Alternative therapeutics like angiotensin receptor blockers have been identified as tentative target candidates but have shown concerns associated with the loss of angiotensin functions crucial for cells [48]. Therefore, to devise effective therapeutics, there is a need to determine the cellular targets in humans that interact with the virus and result in altered functional outcomes. In this study, we uncovered that several human RBPs and miRNAs harbor abundant binding sites across the SARS-CoV-2 genome, illustrating the titration of post-transcriptional regulators. Interestingly, we show that most of these regulators were predominantly expressed in gonadal tissues, adrenal, pancreas, and blood cells.
2. Results and Discussion
We obtained the affinity purification-mass spectrometry (AP-MS)-based SARS-CoV-2 and human proteins interaction network established in HEK293 cells [23] [23] and investigated the human RBPs that directly interact with the viral proteins. Our analysis revealed that SARS-CoV-2-encoded proteins interact directly with 51 human RBPs (Figure 1A). We observed that these primary interacting RBPs were proven to serve several vital functions in the cells, such as polyadenylate binding protein 4 (PABP-4) and Dead-box RNA helicases (DDX21 and DDX10), enzymes involved in translation machinery such as the eukaryotic translation initiation factor 4H (EIF4H), and ribosomal protein L36 (RPL36) (Figure 1A). Among the direct interactors, the highly abundant cytoplasmic PABPs, known to bind the 3′ polyA tail on eukaryotic mRNAs, has previously been reported to interact with polyA tails in bovine coronavirus and the mouse hepatitis virus [49][50][51][49,50,51]. Since SARS-CoV-2 is also composed of polyadenylated RNA, it is likely that the host PABP could modulate the translation of the coronavirus genome through polyA binding. DDX10, another primary interactor observed in the analyzed dataset, has been reported to interact with SARS-CoV-2 nonstructural protein 8 (nsp8) [52], suggesting that the identified host RBPs could be implicated in the regulatory processes of SARS-CoV-2 genome synthesis. EIF4H, also found as one of the primary interactors, was reported to interact with SARS-CoV-2 nonstructural protein 9 (nsp9) in a recently published study [23]. Furthermore, among the immediate interactions, we also found human RBPs such as signal recognition particle 19 (SRP19 and SRP54) and Golgin subfamily B member 1 (GOLGB1) that have been well-recognized for co-translational protein targeting to the membrane and endoplasmic reticulum to Golgi vesicle-mediated transport [53][54] [53,54] (Figure 1A). These results suggest that several human RBPs that come into direct contact with SARS-CoV-2 proteins could contribute to virus assembly and export and could therefore be implicated as therapeutic targets. However, such findings require in-depth experimental validation in a tissue-specific context to support the functional involvement of the identified RBPs in response to SARS-CoV-2 infection.
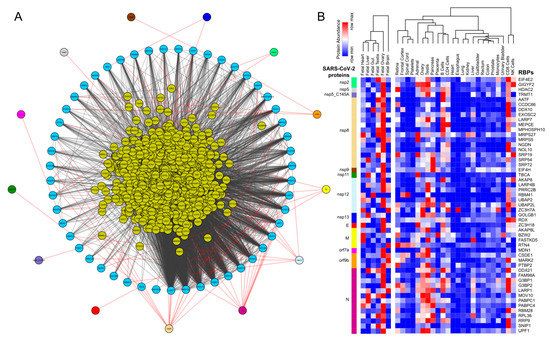
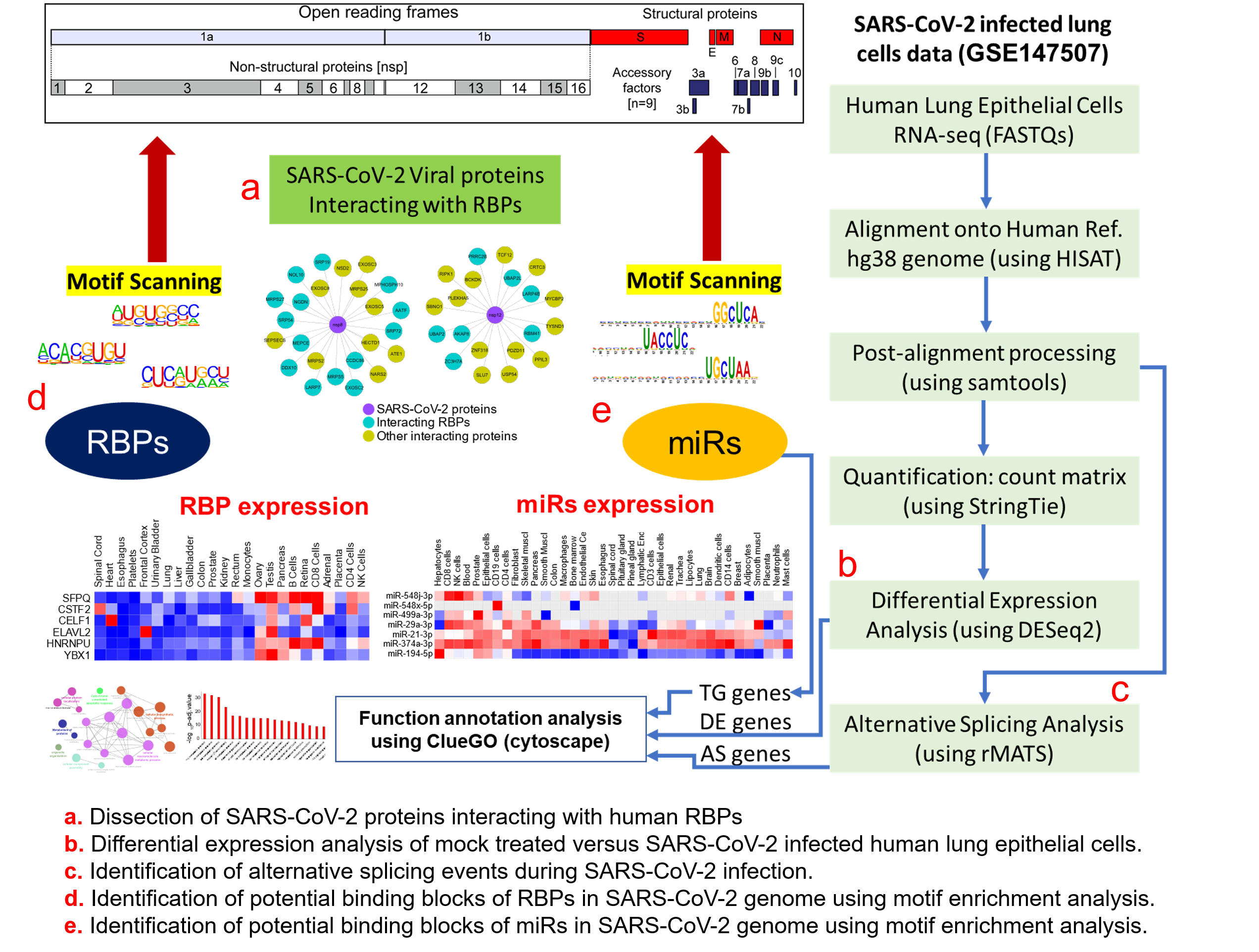
Figure 1. Protein-protein interaction network analysis suggests a direct interaction of human RNA-binding proteins (RBPs) with SARS-CoV-2 viral proteins (A) An integrated SARS-CoV-2—human RBP interaction network. We obtained the mass spectrometry (MS)-based SARS-CoV-2 viral protein to the human protein interaction network established in HEK293 cells and integrated with first-neighbor-interacting RBPs (obtained from BioGRID—https://thebiogrid.org). (B) Protein abundance of SARS-CoV-2-interacting RBPs across human tissues. Expression data was obtained from the human protein map and row normalized. SARS-CoV-2 proteins were color-coded and highlighted in the network.
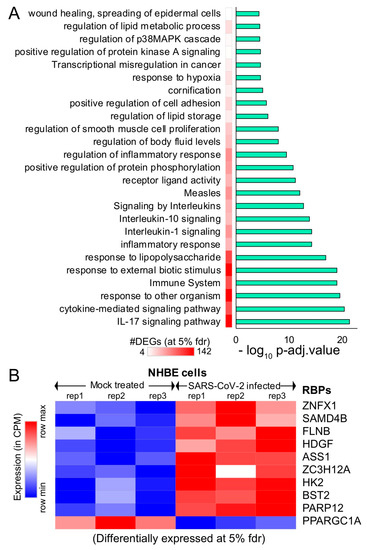
Figure 2. Differential expression analysis of mock-treated vs. SARS-CoV-2-infected primary human lung epithelial cells. (A) Bar plot illustrating the significant pathways obtained from the Gene Ontology (GO) term-based functional grouping of Differentially Expressed Genes (DEGs) at 5% False Discovery Rate (FDR) using ClueGO analysis (Cytoscape plugin) (B) Row normalized expression profile of differentially expressed RBPs in mock-treated and SARS-CoV-2-infected primary human lung epithelial cells (in biological triplicates). NHBE: normal vs. SARS-CoV-2-infected human bronchial epithelial cells.
Figure 3. Alternative splicing events during SARS-CoV-2 infection. (A) Bar plot showing the genes (RBP-encoding genes in blue) exhibiting alternative splicing during SARS-CoV-2 infection in primary human lung epithelial cells (at 5% FDR). (B) Clustered GO term network obtained from the function annotation analysis and grouping of the GO term for the genes exhibiting alternative splicing using ClueGO (Cytoscape plugin). Significant clustering (adj. p < 1 × 10−5) of functional groups were color-coded by functional annotation of the enriched GO biological processes, with the size of the nodes indicating the level of significant association of genes per GO term were shown.
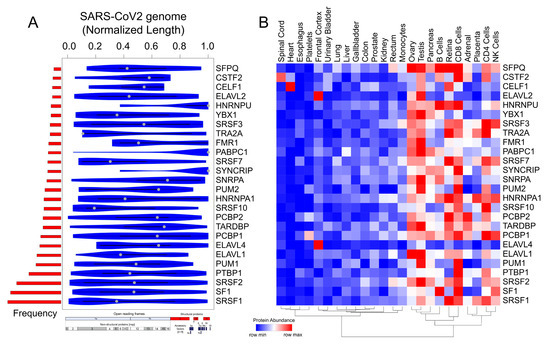
Figure 4. Motif enrichment analysis reveals potential human RBPs titrated by the SARS-CoV-2 viral genome. (A) Violin plot shows the statistically significant (p < 1 × 10−5) preferential binding profile of the RBP motifs (sorted by frequency of binding and greater than 10 sites) across the SARS-CoV-2 viral genome (length normalized) identified using FIMO. (B) Hierarchically clustered heatmap showing the protein abundance (row normalized) of RBPs across tissues.
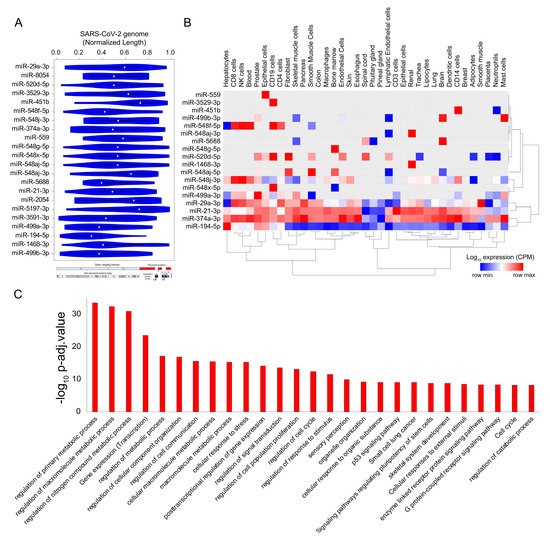
Figure 5. SARS-CoV-2 genome titrates the abundance of functionally important micro-RNAs (miRs) in human tissue. (A) Violin plot shows the statistically significant (p < 1 × 10−5) preferential binding profile of miR motifs (sorted by frequency of binding >15 sites) across the SARS-CoV-2 viral genome (length normalized) identified using FIMO. (B) Hierarchically clustered heatmap showing the log10 expression (Copies Per Million mapped reads (CPM), row normalized) of miRs across the tissues. (C) Bar plot illustrating the significant biological processes obtained from the gene ontology enrichment-based functional grouping of miR target genes (obtained from miRNet). Significant clustering (adj. p < 1 × 10−10) of genes enriched in GO biological processes generated by ClueGO analysis (Cytoscape plugin).