One of the earliest hallmarks of plant immune response is production of reactive oxygen species (ROS) in different subcellular compartments, which regulate plant immunity. A suitable equilibrium, which is crucial to prevent ROS overaccumulation leading to oxidative stress, is maintained by salicylic acid (SA), a chief regulator of ROS. However, ROS not only act downstream of SA signaling, but are also proposed to be a central component of a self-amplifying loop that regulates SA signaling as well as the interaction balance between different phytohormones. The exact role of this crosstalk, the position where SA interferes with ROS signaling and ROS interferes with SA signaling and the outcome of this regulation, depend on the origin of ROS but also on the pathosystem. The precise spatiotemporal regulation of organelle-specific ROS and SA levels determine the effectiveness of pathogen arrest and is therefore crucial for a successful immune response. However, the regulatory interplay behind still remains poorly understood, as up until now, the role of organelle-specific ROS and SA in hypersensitive response (HR)-conferred resistance has mostly been studied by altering the level of a single component. In order to address these aspects, a sophisticated combination of research methods for monitoring the spatiotemporal dynamics of key players and transcriptional activity in plants is needed and will most probably consist of biosensors and precision transcriptomics.
- plant immune response
- reactive oxygen species
- salicylic acid
- reactive oxygen species–salicylic acid crosstalk
- programmed cell death
- hypersensitive-response-conferred resistance
- RBOH NADPH oxidases
- chloroplastic redox state
- biosensors
- precision transcrip
1. Reactive Oxygen Species as One of the Earliest Hallmarks of Plant Immune Response
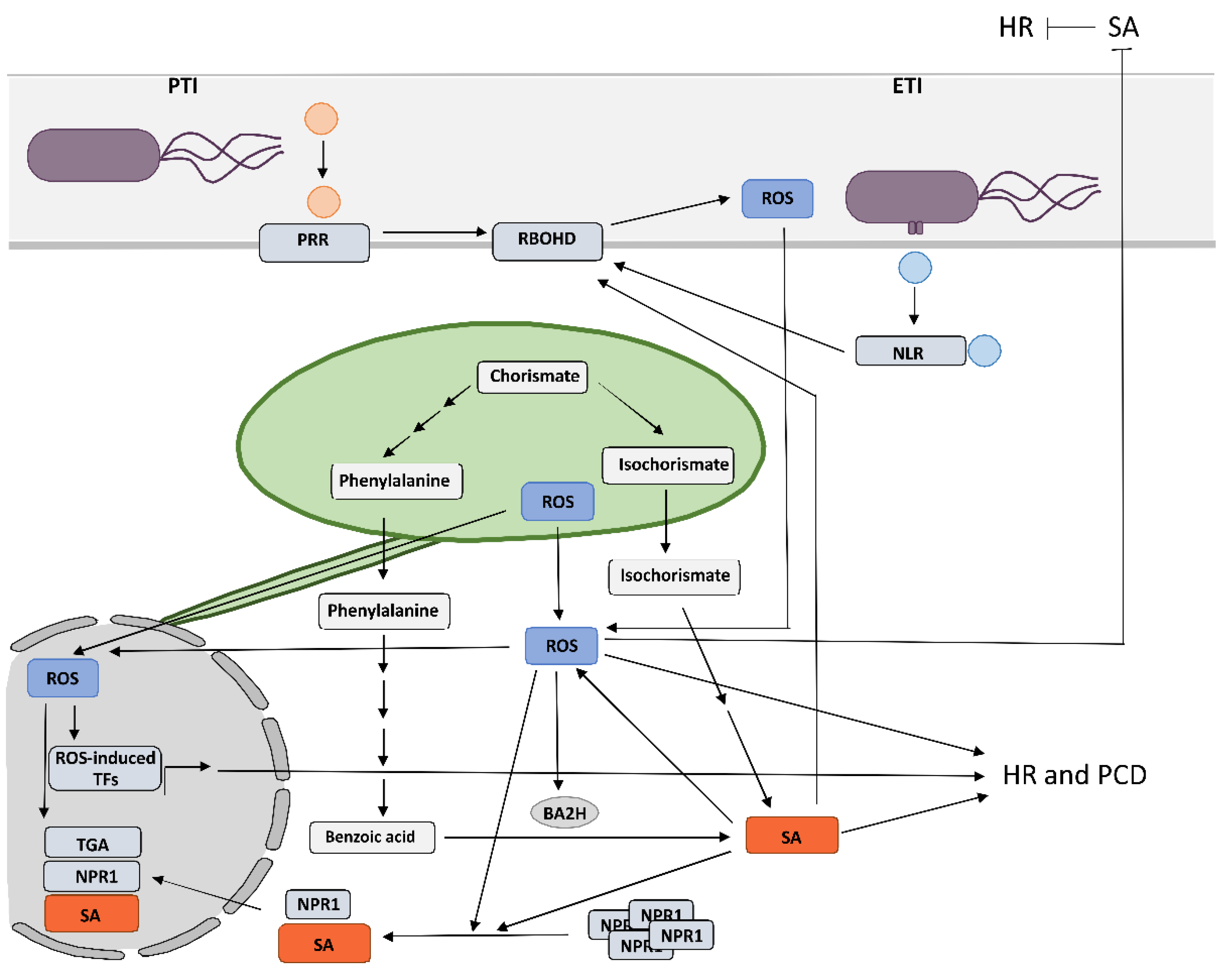
2. Crosstalk between RBOHD-Derived Reactive Oxygen Species and Salicylic Acid in Programmed Cell Death and Resistance
3. Chloroplastic Reactive Oxygen Species Play a Role in the Signaling for Programmed Cell Death and Induce SA-Dependent Transcription of Immune Genes
A growing body of evidence supports a central role of chloroplasts as integrators of environmental signals and key defense organelles, as they host biosynthesis of several key defense-related molecules, including SA and ROS, and are therefore primary sites for the biosynthesis and transmission of pro-defense signals during plant immune responses [7][47][48]. Increases in chloroplastic ROS concentration have been observed in different incompatible plant–pathogen interactions [49], and, in addition, the results of several studies suggest the involvement of chloroplastic ROS in the signaling for and/or execution of HR cell death in HR-conferred resistance [11][50][51][52][53][54][55]. However, its exact role during HR still remains largely elusive. Zurbriggen et al. (2009) suggested that ROS generated in chloroplasts during non-host interaction are essential for the progression of PCD, but do not contribute to the induction of pathogenesis-related genes or other signaling components of the response, including SA signaling [51]. Similarly, Yao and Greenberg did not detect an increase in the expression of genes from SA-mediated signaling in the acd2 mutants that show spontaneous light-dependent PCD and chloroplastic H2O2 increase [56]. In contrast, Straus et al. suggested that chloroplastic ROS acts as a flexible spatiotemporal integration point leading to opposite SA signaling reactions in infected and surrounding tissue to control the propagation of PCD [52]. Predominance of chloroplast superoxide over H2O2 drives PCD in infected tissue and RBOHD-regulated restriction of PCD in the surrounding tissue. When the equilibrium is through SA synthesis shifted towards H2O2 production, this results in runaway PCD [52]. Interestingly, Ochsenbein et al. (2006) also observed that chloroplastic singlet oxygen activates SA-mediated signaling, although SA was not required for a singlet-oxygen-mediated cell death [57]. In potato HR, ROS generated in the chloroplasts around the cell death zone are involved in SA-independent execution of cell death and SA-dependent immune signaling, which are spatially regulated [11][58]. Moreover, recent evidence suggests that chloroplastic ROS might, in addition to signaling in HR cell death, also be involved in controlling plant immune responses by reprogramming transcription of genes involved in response to pathogen attack as one of the retrograde signals [57][59][60][61][62][63][64]. Transmission of pro-defense signals is facilitated by direct connections between chloroplasts and other organelles [65][66]. It has been suggested that stromules, which are extensions in the form of fluid-filled tubules, containing soluble components of the compartments, could facilitate this transmission by enabling more targeted and stronger signal transmission [67]. Stromules are induced by a variety of biotic and abiotic stresses and are involved in retrograde signaling after pathogen invasion, light stress, or movement of chloroplasts within the cell [68][69][70]. However, their direct role in immunity is still largely unresolved [63]. In potato HR-conferred resistance against PVY, stromule formation is induced in close proximity to the cells with oxidized chloroplasts [58]. Since Stonebloom et al. (2012) showed that cell-to-cell transport is negatively regulated by an oxidative shift in chloroplasts, while reductive shift in chloroplasts causes increased cell-to-cell transport, these results could indicate on the potential role of stromules in the signaling for HR-conferred resistance [71]. This is further supported by the fact that stromule formation is induced on the front of virus multiplication zone and is tightly spatiotemporaly regulated by SA signaling [58]. Another type of phenomena that indicate the role of stromules in signaling are the connections of stromules with the plasma membrane, mitochondria, and the nucleus, suggesting that the direct transfer of proteins and metabolites between these organelles and the apoplast could occur [72].4. Reactive Oxygen Species of Different Source and Type Induce Diverse Transcriptional Responses
Distinct subcellular compartments produce different ROS types [73], which could all regulate gene expression [74]. Although each organelle could, in theory, locally manage its own ROS homeostasis, ROS and related signaling intermediates are also involved in interorganellar communication [6][72][75]. For example, chloroplast-derived redox signals could be first integrated in the cytosol or directly transferred to the nucleus, either through physical nucleus–chloroplast interaction or via stromules, in order to control retrograde signaling [76]. Only a small number of proteins targeted to the chloroplast have been also identified in the nucleus to function as retrograde signal transducers in response to biotic and abiotic stresses, some of them being WHIRLY1, the PHD-type transcription factor PTM, and NUCLEAR RECEPTOR INTERACTING PROTEIN 1 (NRIP1) [77]. For example, WHIRLY1 has been proposed to convey the redox status in chloroplasts to the nucleus in a SA-dependent manner [78]. While localization of WHIRLY1 and PTM in both chloroplasts and the nucleus have been shown, the way in which their translocation from chloroplasts to the nucleus occurs is still not known [79][80]. On the other hand, Caplan et al. suggested that after TMV inoculation, the NRIP1 protein is translocated from the chloroplast to the nucleus via stromules [69]. In the nucleus, the control of gene expression depends mainly on the activity of transcription factors (TFs) that interact with oxidative-stress-responsive cis-regulatory elements within the gene promoters. It has been reported that the transcripts generated by increased intracellular H2O2 levels encode proteins of diverse functional categories including TFs, protein kinases, heat shock proteins, glutathione S-transferases (GSTs), UDP-glucuronosyltransferases (UGTs), and cytochrome P450 monooxygenases (CYPs). Upregulated TFs belong to different stress-related TFs’ families, including WRKY, AP2/ERF, MYB, NAC, heat shock factor (HSF), and ZAT [73]. Interestingly, studies have shown that the transcriptional response to apoplastic ROS produced during oxidative burst has little similarity to the effect of chloroplastic ROS [6]. The roles of individual ROS species from different organelles in transcriptional responses have been studied [81], but since the signaling pathways are connected, multiple effects of different organelle-specific ROS on transcriptional response have to be addressed.5. Tools for Studying Redox State with High Spatiotemporal Resolution with Focus on Cytoplasmic and Chloroplastic Redox State in Hypersensitive Response
Despite the new insights that have been brought into the role of redox mechanisms in plant defense response, still one of the major challenges is to monitor local, subcellular, and global ROS dynamics with high selectivity, sensitivity, and spatiotemporal resolution that allow for quantification [82]. Small organic-molecule-based probes have been originally used to measure ROS in plants; however, they have significant limitation as they do not allow for spatiotemporal resolution, since the fluorescence changes as a result of ROS presence are irreversible [83][84]. Nondestructive real-time measuring of redox state in plants with high spatial and temporal resolution is feasible since Jiang et al. reported the use of redox-sensitive green fluorescent protein (roGFP) [85] (Figure 2). Measurement of roGFP fluorescence intensity following excitation with two different wavelengths enables the calculation of the ratio between reduced and oxidized roGFP and thus the determination of the redox state on the cellular or organelle level. The biosensors have been modified by the addition of signal sequences to target them to different subcellular organelles, while two variants, roGFP1 and roGFP2, with different excitation and emission spectra, were developed to allow for optimal selection according to the redox state in the particular organelle. By fusing the peroxisomal targeting peptide sequence SKL, per-roGFP1 and px-roGFP2 were targeted to peroxisomes [86][87]. mt-roGFP1 and mt-roGFP2 are available for measuring the redox state in mitochondria due to the fusion with mitochondrial localization signal peptide from the tobacco b-ATPase [85][87], while er-roGFP2 was constructed by fusing roGFP2 with the endoplasmic reticulum (ER) retention signal peptide HDEL for following the redox state in ER [88]. Finally, cp-GFP2, pt-roGFP2, and chl-roGFP2 were constructed by adding plastid-targeting signal peptide TKTP, coding sequence for RuBisCo small subunit transit peptide, or the first 74 amino acids from PRXa to roGFP2 coding sequence, respectively, for measuring change of redox state in chloroplasts [71][89][90]. Since pt-roGFP is targeted to the chloroplast outer plastid envelope membrane, it also allows for the imaging of stromules formation [68]. The above-mentioned sensors were used in tobacco, Arabidopsis, and potato subjected to different developmental and environmental stresses [68][71][85][86][87][88][89][90][91][92][93][94], some of them also in HR [58][95][96]. roGFP2 expressed in the cytosol senses the redox potential of the cellular glutathione buffer via glutaredoxin (GRX) as a mediator of reversible electron flow between glutathione and roGFP2 [88]. To facilitate specific real-time equilibration between roGFP2 and the glutathione redox couple, fusion constructs with human glutaredoxin 1 (GRX1) Grx1-roGFP2 and roGFP2-Grx2 were generated [97][98][99]. Moreover, two roGFP derivatives, roGFP2-iL and roGFP1-iX, with different midpoint potential and excitation properties, were developed to further extend the range of suitable probes [100]. Another group of genetically encoded sensors that detect H2O2 levels instead of measuring glutathione redox potential was also developed. roGFP2-Orp1, based on a redox relay between the GPX-like enzyme oxidant receptor peroxidase-1 (Orp1) from Saccharomyces cerevisiae and roGFP2 was generated for sensing transient changes in H2O2 [93]. Another sensor that reports on local alterations in H2O2 concentrations exploits the H2O2-sensitive bacterial transcription factor OxyR for its response and was named HyPer [101][102]. The above-mentioned types of sensors were also used to follow redox potential and H2O2 in the cytoplasm during HR [103][104][105][106][107][108] or were further upgraded to follow redox potential in different cellular compartments, including chloroplasts [109]. Using HyPer2, Exposito-Rodriguez et al. showed that in photosynthetic Nicotiana benthamiana epidermal cells, exposure to high light increased H2O2 production in chloroplast stroma, cytosol, and nuclei, suggesting direct H2O2 transfer from chloroplasts to nuclei [110]. Other genetically encoded redox and H2O2 sensors are summareviewized in [82][111]. The most recent ones are biosensor CROST for measurements of the thioredoxin redox state in chloroplasts [112], FRET-based biosensors, and ROS regulated promoter–FP fusions [82][113].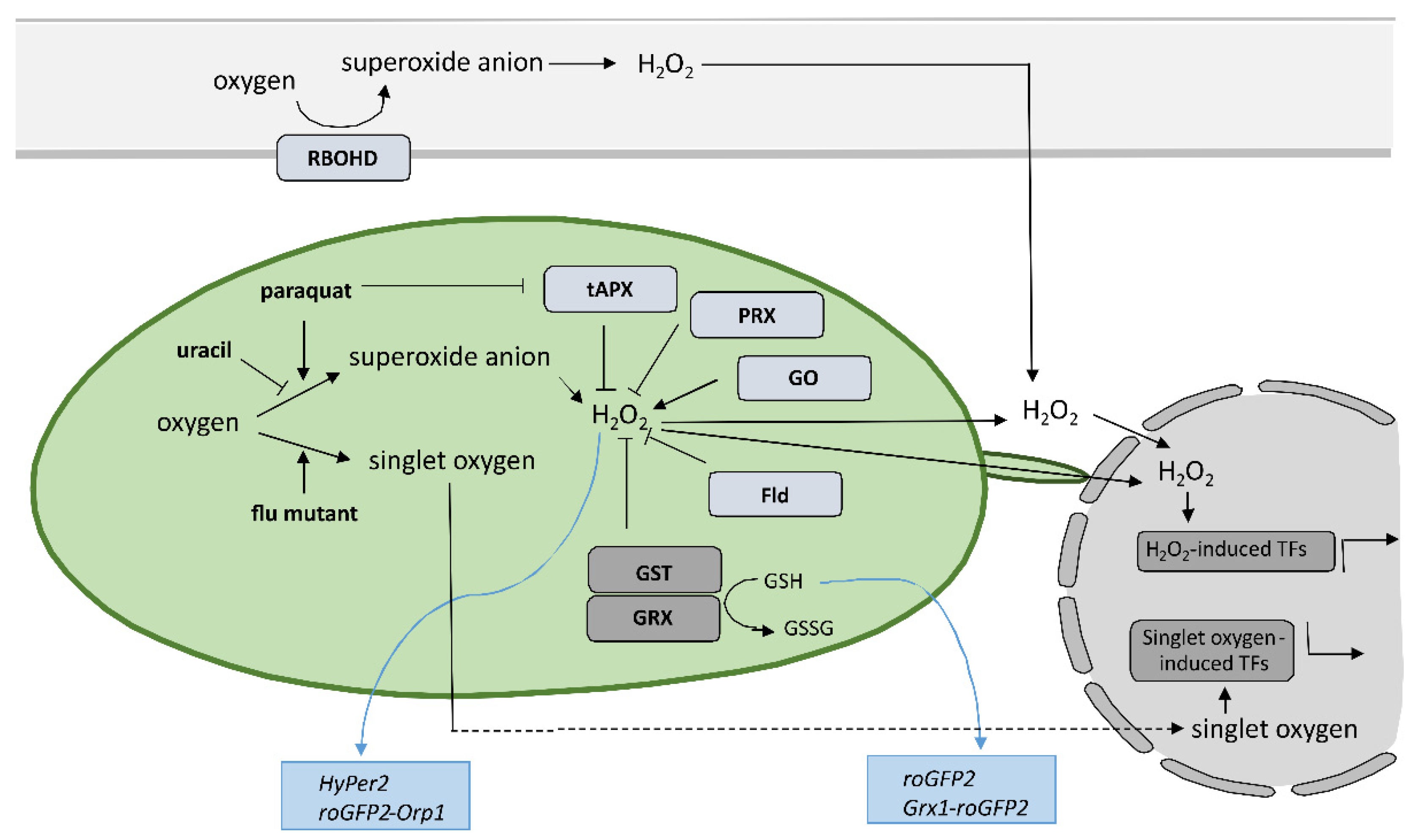
6. Conclusions
The results of the above-mentioned studies suggest that the precise spatiotemporal regulation of key players, including organelle-specific ROS and SA levels, determines the effectiveness of pathogen arrest and is therefore crucial for a successful immune response. The change of SA and ROS levels and other key players alter the rate of cell-to-cell and systemic pathogen spread, rate of cell death induction, and spatial transcriptional response, leading to susceptibility or resistance. WeScholars suggest that only a coordinated and intertwined action of all main components enable effective immune response. However, the specific interactions between them and the regulatory interplay behind still remain poorly understood, as up until now, the role of organelle-specific ROS and SA in HR-conferred resistance has only been studied by altering the level of a single component. In order to address these aspects, a sophisticated combination of research methods for monitoring the spatiotemporal dynamics of key players and transcriptional activity in plants is needed. The precise sampling of tissue sections surrounding the HR-PCD, with spatial resolution and suitable for transcriptomics analyses [11], in combination with the use of biosensors [130], could enable identification of novel key players and could unravel the interconnectivity of immune signaling components. Such an approach could therefore present a step forward in studying the resistance response.
References
- Lu, Y.; Tsuda, K. Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol. Plant-Microbe Interact. 2021, 34, 3–14.
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329.
- Künstler, A.; Bacsó, R.; Gullner, G.; Hafez, Y.M.; Király, L. Staying alive—Is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 2016, 93, 75–84.
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178.
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412.
- Shapiguzov, A.; Julia, P.; Wrzaczek, M.; Kangasjärvi, J. ROS-talk—How the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012, 3, 292.
- Lu, Y.; Yao, J. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci. 2018, 19, 3900.
- Kadota, Y.; Liebrand, T.W.H.; Goto, Y.; Sklenar, J.; Derbyshire, P.; Menke, F.L.H.; Torres, M.; Molina, A.; Zipfel, C.; Coaker, G.; et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol. 2019, 221, 2160–2175.
- Hu, C.H.; Wang, P.Q.; Zhang, P.P.; Nie, X.M.; Li, B.B.; Tai, L.; Liu, W.T.; Li, W.Q.; Chen, K.M. NADPH oxidases: The vital performers and center hubs during plant growth and signaling. Cells 2020, 9, 437.
- Hernández, J.A.; Gullner, G.; Clemente-Moreno, M.J.; Künstler, A.; Juhász, C.; Díaz-Vivancos, P.; Király, L. Oxidative stress and antioxidative responses in plant–virus interactions. Physiol. Mol. Plant Pathol. 2016, 94, 134–148.
- Lukan, T.; Pompe-Novak, M.; Baebler, Š.; Tušek-Žnidarič, M.; Kladnik, A.; Križnik, M.; Blejec, A.; Zagorščak, M.; Stare, K.; Dušak, B.; et al. Precision transcriptomics of viral foci reveals the spatial regulation of immune-signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. Plant J. 2020, 104, 645–661.
- Mur, L.A.J.; Bi, Y.; Darby, R.M.; Firek, S.; Draper, J. Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J. 1997, 12, 1113–1126.
- Liao, Y.; Tian, M.; Zhang, H.; Li, X.; Wang, Y.; Xia, X.; Zhou, J.; Zhou, Y.; Yu, J.; Shi, K.; et al. Salicylic acid binding of mitochondrial alpha-ketoglutarate dehydrogenase E2 affects mitochondrial oxidative phosphorylation and electron transport chain components and plays a role in basal defense against tobacco mosaic virus in tomato. New Phytol. 2015, 205, 1296–1307.
- Torres, M.A.; Dangl, J.L.; Jones, J.D.G. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522.
- Chaouch, S.; Queval, G.; Noctor, G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intra-cellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012, 69, 613–627.
- Liu, H.B.; Wang, X.D.; Zhang, Y.Y.; Dong, J.J.; Ma, C.; Chen, W.L. NADPH oxidase RBOHD contributes to autophagy and hypersensitive cell death during the plant defense response in Arabidopsis thaliana. Biol. Plant. 2015, 59, 570–580.
- Yoshie, Y.; Goto, K.; Takai, R.; Iwano, M.; Takayama, S.; Isogai, A.; Che, F.S. Function of the rice gp91phox homologs OsrbohA and OsrbohE genes in ROS-dependent plant immune responses. Plant Biotechnol. 2005, 22, 127–135.
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 2016, 67, 1663–1676.
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.G.; Doke, N. Nicotiana benthamiana gp91 phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 2003, 15, 706–718.
- Zhang, Z.; van Esse, H.P.; van Damme, M.; Fradin, E.F.; Liu, C.M.; Thomma, B.P.H.J. Ve1-mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol. Plant Pathol. 2013, 14, 719–727.
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Pathogen-induced, NADPH oxidase–derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134.
- Berrocal-Lobo, M.; Stone, S.; Yang, X.; Antico, J.; Callis, J.; Ramonell, K.M. ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase- mediated defense responses. PLoS ONE 2010, 5, e14426.
- Proels, R.K.; Oberhollenzer, K.; Pathuri, I.P.; Hensel, G.; Kumlehn, J.; Hückelhoven, R.; Phytopathologie, L.; München, T.U. RBOHF2 of Barley Is required for normal development of penetration resistance to the parasitic fungus Blumeria graminis f. sp. hordei. Mol. Plant-Microbe Interact. 2010, 23, 1143–1150.
- Yao, Z.; Islam, M.R.; Badawi, M.A.; El-Bebany, A.F.; Daayf, F. Overexpression of StRbohA in Arabidopsis thaliana enhances defence responses against Verticillium dahliae. Physiol. Mol. Plant Pathol. 2015, 90, 105–114.
- Foley, R.C.; Gleason, C.A.; Anderson, J.P.; Hamann, T.; Singh, K.B. Genetic and genomic analysis of rhizoctonia solani interactions with arabidopsis; evidence of resistance mediated through NADPH oxidases. PLoS ONE 2013, 8, e56814.
- Trujillo, M.; Altschmied, L.; Schweizer, P.; Kogel, K.H.; Hückelhoven, R. Respiratory burst oxidase homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp. hordei. J. Exp. Bot. 2006, 57, 3781–3791.
- Pieterse, C.M.J.; van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521.
- Huang, W.; Wang, Y.; Li, X.; Zhang, Y. Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant immunity. Mol. Plant 2020, 13, 31–41.
- Maruri-López, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and extracellular journey of the phytohormone salicylic acid. Front. Plant Sci. 2019, 10, 423.
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36.
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800.
- Saleem, M.; Fariduddin, Q.; Castroverde, C.D.M. Salicylic acid: A key regulator of redox signalling and plant immunity. Plant Physiol. Biochem. 2021, 168, 381–397.
- Chen, Z.; Silva, H.; Klessig, D.F. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 1993, 262, 1883–1886.
- Mur, L.A.J.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 2008, 59, 501–520.
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, 711–723.
- Chivasa, S.; Murphy, A.M.; Naylor, M.; Carr, J.P. Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 1997, 9, 547–557.
- Chivasa, S.; Carr, J.P. Cyanide restores N gene-mediated resistance to tobacco mosaic virus in transgenic tobacco expressing salicylic acid hydroxylase. Plant Cell 1998, 10, 1489–1498.
- Baebler, Š.; Witek, K.; Petek, M.; Stare, K.; Tušek-Znidaric, M.; Pompe-Novak, M.; Renaut, J.; Szajko, K.; Strzelczyk-Zyta, D.; Marczewski, W.; et al. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against potato virus Y infection in potato. J. Exp. Bot. 2014, 65, 1095–1109.
- Liu, Y.; He, C. Regulation of plant reactive oxygen species (ROS) in stress responses: Learning from AtRBOHD. Plant Cell Rep. 2016, 35, 995–1007.
- Yi, S.Y.; Shirasu, K.; Moon, J.S.; Lee, S.G.; Kwon, S.Y. The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS ONE 2014, 9, e88951.
- Chang, Y.L.; Li, W.Y.; Miao, H.; Yang, S.Q.; Li, R.; Wang, X.; Li, W.Q.; Chen, K.M. Comprehensive genomic analysis and expression profiling of the NOX gene families under abiotic stresses and hormones in plants. Genome Biol. Evol. 2016, 8, 791–810.
- Kaur, G.; Pati, P.K. Analysis of cis-acting regulatory elements of Respiratory burst oxidase homolog (Rboh) gene families in Arabidopsis and rice provides clues for their diverse functions. Comput. Biol. Chem. 2016, 62, 104–118.
- Yoshioka, H.; Sugie, K.; Park, H.J.; Maeda, H.; Tsuda, N.; Kawakita, K.; Doke, N. Induction of plant gp91 phox homolog by fungal cell wall, arachidonic acid, and salicylic acid in potato. Mol. Plant-Microbe Interact. 2001, 14, 725–736.
- Xi, C.; Guohui, L.; Muhammad Aamir, M.; Han, W.; Muhammad, A.; Xueqiang, S.; Jingyun, Z.; Taoshan, J.; Qing, J.; Yongping, C.; et al. In silico genome-wide analysis of respiratory burst oxidase homolog (RBOH) family genes in five fruit-producing trees, and potential functional analysis on lignification of stone cells in Chinese white pear. Cells 2019, 8, 520.
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429.
- Pogány, M.; von Rad, U.; Grün, S.; Dongó, A.; Pintye, A.; Simoneau, P.; Bahnweg, G.; Kiss, L.; Barna, B.; Durner, J. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 2009, 151, 1459–1475.
- Serrano, I.; Audran, C.; Rivas, S. Chloroplasts at work during plant innate immunity. J. Exp. Bot. 2016, 67, 3845–3854.
- Schwenkert, S.; Fernie, A.R.; Geigenberger, P.; Leister, D.; Möhlmann, T.; Naranjo, B.; Neuhaus, H.E. Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci. 2022, 27, 577–587.
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.R. ROS signaling in the hypersensitive response: When, where and what for? Plant Signal. Behav. 2010, 5, 393–396.
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S.; Life, B. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007, 51, 941–954.
- Zurbriggen, M.D.; Carrillo, N.; Tognetti, V.B.; Melzer, M.; Peisker, M.; Hause, B.; Hajirezaei, M.R. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. Plant J. 2009, 60, 962–973.
- Straus, M.R.; Rietz, S.; ver Loren van Themaat, E.; Bartsch, M.; Parker, J.E. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 2010, 62, 628–640.
- Ishiga, Y.; Ishiga, T.; Wangdi, T.; Mysore, K.S.; Uppalapati, S.R. NTRC and chloroplast-generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis. Mol. Plant-Microbe Interact. 2012, 25, 294–306.
- Kim, C.; Meskauskiene, R.; Zhang, S.; Lee, K.P.; Ashok, M.L.; Blajecka, K.; Herrfurth, C.; Feussner, I.; Apela, K. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 2012, 24, 3026–3039.
- Xu, Q.; Tang, C.; Wang, X.; Sun, S.; Zhao, J.; Kang, Z.; Wang, X. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 2019, 10, 5571.
- Yao, N.; Greenberg, J.T. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 2006, 18, 397–411.
- Ochsenbein, C.; Przybyla, D.; Danon, A.; Landgraf, F.; Göbel, C.; Imboden, A.; Feussner, I.; Apel, K. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J. 2006, 47, 445–456.
- Lukan, T.; Županič, A.; Mahkovec Povalej, T.; Brunkard, J.O.; Juteršek, M.; Baebler, Š.; Gruden, K. Chloroplast redox state changes indicate cell-to-cell signalling during the hypersensitive response. bioRxiv 2021.
- Lee, K.P.; Kim, C.; Landgraf, F.; Apel, K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 10270–10275.
- Noshi, M.; Maruta, T.; Shigeoka, S. Relationship between chloroplastic H2O2 and the salicylic acid response. Plant Signal. Behav. 2012, 7, 944–946.
- Maruta, T.; Noshi, M.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 2012, 287, 11717–11729.
- Nomura, H.; Komori, T.; Uemura, S.; Kanda, Y.; Shimotani, K.; Nakai, K.; Furuichi, T.; Takebayashi, K.; Sugimoto, T.; Sano, S.; et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 2012, 3, 910–926.
- Sewelam, N.; Jaspert, N.; van der Kelen, K.; Tognetti, V.B.; Schmitz, J.; Frerigmann, H.; Stahl, E.; Zeier, J.; van Breusegem, F.; Maurino, V.G. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 2014, 7, 1191–1210.
- Pierella Karlusich, J.J.; Zurbriggen, M.D.; Shahinnia, F.; Sonnewald, S.; Sonnewald, U.; Hosseini, S.A.; Hajirezaei, M.R.; Carrillo, N. Chloroplast redox status modulates genome-wide plant responses during the non-host interaction of Tobacco with the hemibiotrophic bacterium Xanthomonas campestris pv. vesicatoria. Front. Plant Sci. 2017, 8, 1158.
- Kmiecik, P.; Leonardelli, M.; Teige, M. Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 2016, 67, 3793–3807.
- Pérez-Sancho, J.; Tilsner, J.; Samuels, A.L.; Botella, M.A.; Bayer, E.M.; Rosado, A. Stitching organelles: Organization and function of specialized membrane contact sites in plants. Trends Cell Biol. 2016, 26, 705–717.
- Hanson, M.R.; Conklin, P.L. Stromules, functional extensions of plastids within the plant cell. Curr. Opin. Plant Biol. 2020, 58, 25–32.
- Brunkard, J.O.; Runkel, A.M.; Zambryski, P.C. Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. USA 2015, 112, 10044–10049.
- Caplan, J.L.; Kumar, A.S.; Park, E.; Padmanabhan, M.S.; Hoban, K.; Modla, S.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplast stromules function during innate immunity. Dev. Cell 2015, 34, 45–57.
- Kumar, A.S.; Park, E.; Nedo, A.; Alqarni, A.; Ren, L.; Hoban, K.; Modla, S.; McDonald, J.H.; Kambhamettu, C.; Dinesh-Kumar, S.P.; et al. Stromule extension along microtubules coordinated with actin-mediated anchoring guides perinuclear chloroplast movement during innate immunity. eLife 2018, 7, e23625.
- Stonebloom, S.; Brunkard, J.O.; Cheung, A.C.; Jiang, K.; Feldman, L.; Zambryski, P. Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol. 2012, 158, 190–199.
- Noctor, G.; Foyer, C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016, 171, 1581–1592.
- He, H.; van Breusegem, F.; Mhamdi, A. Redox-dependent control of nuclear transcription in plants. J. Exp. Bot. 2018, 69, 3359–3372.
- Locato, V.; Cimini, S.; de Gara, L. ROS and redox balance as multifaceted players of cross-tolerance: Epigenetic and retrograde control of gene expression. J. Exp. Bot. 2018, 69, 3373–3391.
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844.
- Bleau, J.R.; Spoel, S.H. Selective redox signaling shapes plant-pathogen interactions. Plant Physiol. 2021, 186, 53–65.
- Mullineaux, P.M.; Exposito-Rodriguez, M.; Laissue, P.P.; Smirnoff, N.; Park, E. Spatial chloroplast-to-nucleus signalling involving plastid–nuclear complexes and stromules. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190405.
- Foyer, C.H.; Karpinska, B.; Krupinska, K. The functions of WHIRLY1 and REDOXRESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: A hypothesis. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130226.
- Grabowski, E.; Miao, Y.; Mulisch, M.; Krupinska, K. Single-stranded DNA-binding protein whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 2008, 147, 1800–1804.
- Sun, X.; Feng, P.; Xu, X.; Guo, H.; Ma, J.; Chi, W.; Lin, R.; Lu, C.; Zhang, L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2011, 2, 477.
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Saifullah; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369.
- Ortega-Villasante, C.; Burén, S.; Blázquez-Castro, A.; Barón-Sola, Á.; Hernández, L.E. Fluorescent in vivo imaging of reactive oxygen species and redox potential in plants. Free Radic. Biol. Med. 2018, 122, 202–220.
- Choi, W.G.; Swanson, S.J.; Gilroy, S. High-resolution imaging of Ca 2+, redox status, ROS and pH using GFP biosensors. Plant J. 2012, 70, 118–128.
- Fichman, Y.; Miller, G.; Mittler, R. Whole-plant live imaging of reactive oxygen species. Mol. Plant 2019, 12, 1203–1210.
- Jiang, K.; Schwarzer, C.; Lally, E.; Zhang, S.; Ruzin, S.; Machen, T.; Remington, S.J.; Feldman, L. Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 2006, 141, 397–403.
- Rosenwasser, S.; Rot, I.; Meyer, A.J.; Feldman, L.; Jiang, K.; Friedman, H. A fluorometer-based method for monitoring oxidation of redox-sensitive GFP (roGFP) during development and extended dark stress. Physiol. Plant. 2010, 138, 493–502.
- Schwarzländer, M.; Fricker, M.D.; Müller, C.; Marty, L.; Brach, T.; Novak, J.; Sweetlove, L.J.; Hell, R.; Meyer, A.J. Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 2008, 231, 299–316.
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.P.; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007, 52, 973–986.
- Schwarzländer, M.; Fricker, M.D.; Sweetlove, L.J. Monitoring the in vivo redox state of plant mitochondria: Effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim. Biophys. Acta-Bioenerg. 2009, 1787, 468–475.
- Haber, Z.; Lampl, N.; Meyer, A.J.; Zelinger, E.; Hipsch, M.; Rosenwasser, S. Resolving diurnal dynamics of the chloroplastic glutathione redox state in Arabidopsis reveals its photosynthetically derived oxidation. Plant Cell 2021, 33, 1828–1844.
- Kidd, B.N.; Foley, R.; Singh, K.B.; Anderson, J.P. Foliar resistance to Rhizoctonia solani in Arabidopsis is compromised by simultaneous loss of ethylene, jasmonate and PEN2 mediated defense pathways. Sci. Rep. 2021, 11, 2546.
- Hipsch, M.; Lampl, N.; Zelinger, E.; Barda, O.; Waiger, D.; Rosenwasser, S. Sensing stress responses in potato with whole-plant redox imaging. Plant Physiol. 2021, 187, 618–631.
- Nietzel, T.; Elsässer, M.; Ruberti, C.; Steinbeck, J.; Ugalde, J.M.; Fuchs, P.; Wagner, S.; Ostermann, L.; Moseler, A.; Lemke, P.; et al. The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2O2 and thiol redox integration and elucidates intracellular H2O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. New Phytol. 2019, 221, 1649–1664.
- Fuchs, R.; Kopischke, M.; Klapprodt, C.; Hause, G.; Meyer, A.J.; Schwarzländer, M.; Fricker, M.D.; Lipka, V. Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in arabidopsis. Plant Cell 2016, 28, 130–145.
- Maughan, S.C.; Pasternak, M.; Cairns, N.; Kiddle, G.; Brach, T.; Jarvis, R.; Haas, F.; Nieuwland, J.; Lim, B.; Müller, C.; et al. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2331–2336.
- Krönauer, C.; Lahaye, T. The flavin monooxygenase Bs3 triggers cell death in plants, impairs growth in yeast and produces H2O2 in vitro. PLoS ONE 2021, 16, e0256217.
- Gutscher, M.; Pauleau, A.L.; Marty, L.; Brach, T.; Wabnitz, G.H.; Samstag, Y.; Meyer, A.J.; Dick, T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 2008, 5, 553–559.
- Marty, L.; Siala, W.; Schwarzländer, M.; Fricker, M.D.; Wirtz, M.; Sweetlove, L.J.; Meyer, Y.; Meyer, A.J.; Reichheld, J.P.; Hell, R. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 9109–9114.
- Albrecht, S.C.; Sobotta, M.C.; Bausewein, D.; Aller, I.; Hell, R.; Dick, T.P.; Meyer, A.J. Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J. Biomol. Screen. 2014, 19, 379–386.
- Aller, I.; Rouhier, N.; Meyer, A.J. Development of roGFP2-derived redox probes for measurement of the glutathione redox potential in the cytosol of severely glutathione-deficient rml1 seedlings. Front. Plant Sci. 2013, 4, 506.
- Costa, A.; Drago, I.; Behera, S.; Zottini, M.; Pizzo, P.; Schroeder, J.I.; Pozzan, T.; Schiavo, F. Lo H2O2 in plant peroxisomes: An in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 2010, 62, 760–772.
- Ugalde, J.M.; Schlößer, M.; Dongois, A.; Martinière, A.; Meyer, A.J. The latest HyPe(r) in plant H2O2 biosensing. Plant Physiol. 2021, 187, 480–484.
- Dubreuil-Maurizi, C.; Poinssot, B. Role of glutathione in plant signaling under biotic stress. Plant Signal. Behav. 2012, 7, 210–212.
- Dubreuil-Maurizi, C.; Vitecek, J.; Marty, L.; Branciard, L.; Frettinger, P.; Wendehenne, D.; Meyer, A.J.; Mauch, F.; Poinssot, B. Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 2011, 157, 2000–2012.
- Matern, S.; Peskan-Berghoefer, T.; Gromes, R.; Kiesel, R.V.; Rausch, T. Imposed glutathione-mediated redox switch modulates the tobacco wound-induced protein kinase and salicylic acid-induced protein kinase activation state and impacts on defence against Pseudomonas syringae. J. Exp. Bot. 2015, 66, 1935–1950.
- Hussain, J.; Chen, J.; Locato, V.; Sabetta, W.; Behera, S.; Cimini, S.; Griggio, F.; Martínez-Jaime, S.; Graf, A.; Bouneb, M.; et al. Constitutive cyclic GMP accumulation in Arabidopsis thaliana compromises systemic acquired resistance induced by an avirulent pathogen by modulating local signals. Sci. Rep. 2016, 6, 36423.
- Doccula, F.G.; Luoni, L.; Behera, S.; Bonza, M.C.; Costa, A. In vivo analysis of calcium levels and glutathione redox status in Arabidopsis epidermal leaf cells infected with the hypersensitive response-inducing bacteria pseudomonas. Methods Mol. Biol. 2018, 1743, 125–141.
- Mencia, R.; Céccoli, G.; Fabro, G.; Torti, P.; Colombatti, F.; Ludwig-Müller, J.; Alvarez, M.E.; Welchen, E. OXR2 increases plant defense against a hemibiotrophic pathogen via the salicylic acid pathway. Plant Physiol. 2020, 184, 1112–1127.
- Ugalde, J.M.; Fuchs, P.; Nietzel, T.; Cutolo, E.A.; Homagk, M.; Vothknecht, U.C.; Holuigue, L.; Schwarzländer, M.; Müller-Schüssele, S.J.; Meyer, A.J. Chloroplast-derived photo-oxidative stress causes changes in H2O2 and EGSH in other subcellular compartments. Plant Physiol. 2021, 186, 125–141.
- Exposito-Rodriguez, M.; Laissue, P.P.; Yvon-Durocher, G.; Smirnoff, N.; Mullineaux, P.M. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 2017, 8, 4.
- Kostyuk, A.I.; Panova, A.S.; Kokova, A.D.; Kotova, D.A.; Maltsev, D.I.; Podgorny, O.V.; Belousov, V.V.; Bilan, D.S. In vivo imaging with genetically encoded redox biosensors. Int. J. Mol. Sci. 2020, 21, 8164.
- Sugiura, K.; Yokochi, Y.; Fu, N.; Fukaya, Y.; Yoshida, K.; Mihara, S.; Hisabori, T. The thioredoxin (Trx) redox state sensor protein can visualize Trx activities in the light/dark response in chloroplasts. J. Biol. Chem. 2019, 294, 12091–12098.
- Ortega-Villasante, C.; Burén, S.; Barón-Sola, Á.; Martínez, F.; Hernández, L.E. In vivo ROS and redox potential fluorescent detection in plants: Present approaches and future perspectives. Methods 2016, 109, 92–104.
- Dogra, V.; Kim, C. Singlet oxygen metabolism: From genesis to signaling. Front. Plant Sci. 2020, 10, 1640.
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Biol. 1999, 50, 601–639.
- Awad, J.; Stotz, H.U.; Fekete, A.; Krischke, M.; Engert, C.; Havaux, M.; Berger, S.; Mueller, M.J. 2-cysteine peroxiredoxins and thylakoid ascorbate peroxidase create awater-water cycle that is essential to protect the photosynthetic apparatus under high light stress conditions. Plant Physiol. 2015, 167, 1592–1603.
- Murgia, I.; Tarantino, D.; Vannini, C.; Bracale, M.; Carravieri, S.; Soave, C. Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J. 2004, 38, 940–953.
- Tognetti, V.B.; Palatnik, J.F.; Fillat, M.F.; Melzer, M.; Hajirezaei, M.R.; Valle, E.M.; Carrillo, N. Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. Plant Cell 2006, 18, 2035–2050.
- Tognetti, V.B.; Zurbriggen, M.D.; Morandi, E.N.; Fillat, M.F.; Valle, E.M.; Hajirezaei, M.R.; Carrillo, N. Enhanced plant tolerance to iron starvation by functional substitution of chloroplast ferredoxin with a bacterial flavodoxin. Proc. Natl. Acad. Sci. USA 2007, 104, 11495–11500.
- Zurbriggen, M.D.; Tognetti, V.B.; Fillat, M.F.; Hajirezaei, M.R.; Valle, E.M.; Carrillo, N. Combating stress with flavodoxin: A promising route for crop improvement. Trends Biotechnol. 2008, 26, 531–537.
- Coba de la Peña, T.; Redondo, F.J.; Manrique, E.; Lucas, M.M.; Pueyo, J.J. Nitrogen fixation persists under conditions of salt stress in transgenic Medicago truncatula plants expressing a cyanobacterial flavodoxin. Plant Biotechnol. J. 2010, 8, 954–965.
- Li, Z.; Yuan, S.; Jia, H.; Gao, F.; Zhou, M.; Yuan, N.; Wu, P.; Hu, Q.; Sun, D.; Luo, H. Ectopic expression of a cyanobacterial flavodoxin in creeping bentgrass impacts plant development and confers broad abiotic stress tolerance. Plant Biotechnol. J. 2017, 15, 433–446.
- Pierella Karlusich, J.J.; Arce, R.C.; Shahinnia, F.; Sonnewald, S.; Sonnewald, U.; Zurbriggen, M.D.; Hajirezaei, M.R.; Carrillo, N. Transcriptional and metabolic profiling of potato plants expressing a plastid-targeted electron shuttle reveal modulation of genes associated to drought tolerance by chloroplast redox poise. Int. J. Mol. Sci. 2020, 21, 7199.
- Fahnenstich, H.; Scarpeci, T.E.; Valle, E.M.; Flügge, U.I.; Maurino, V.G. Generation of hydrogen peroxide in chloroplasts of arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 2008, 148, 719–729.
- Schmidt, A.; Mächtel, R.; Ammon, A.; Engelsdorf, T.; Schmitz, J.; Maurino, V.G.; Voll, L.M. Reactive oxygen species dosage in Arabidopsis chloroplasts can improve resistance towards Colletotrichum higginsianum by the induction of WRKY33. New Phytol. 2020, 226, 189–204.
- Meskauskiene, R.; Nater, M.; Goslings, D.; Kessler, F.; Op den Camp, R.; Apel, K. FLU: A negative regulator of chlorophyll biosynthesis in arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 12826–12831.
- Scarpeci, T.E.; Zanor, M.I.; Carrillo, N.; Mueller-Roeber, B.; Valle, E.M. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: A focus on rapidly induced genes. Plant Mol. Biol. 2008, 66, 361–378.
- Mano, J.; Ohno, C.; Domae, Y.; Asada, K. Chloroplastic ascorbate peroxidase is the primary target of methylviologen-induced photooxidative stress in spinach leaves: Its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim. Biophys. Acta—Bioenerg. 2001, 1504, 275–287.
- Abdollahi, H.; Ghahremani, Z. The role of chloroplasts in the interaction between Erwinia amylovora and host plants. Acta Hortic. 2011, 896, 215–222.
- Levak, V.; Lukan, T.; Gruden, K.; Coll, A. Biosensors: A sneak peek into plant cell’s immunity. Life 2021, 11, 209.
