The entreviewy talks about Pepper vein banding virus (PVBV) and its encoded proteins, particularly focusing on the polyprotein processing and its regulation. Moreover, PVBV as a nanoparticle and its potential applications in immunodiagnostics and therapeutics has also been discussed,.
- Potyvirus, VLPs, intrinsically disordered proteins
Note:All the information in this draft can be edited by authors. And the entry will be online only after authors edit and submit it.
Definition: Pepper vein banding virus (PVBV) is a distinct species in the Potyvirus genus which infects economically important plants in several parts of India. Like other potyviruses, PVBV encodes multifunctional proteins, with several interaction partners, having implications at different stages of the potyviral infection.
1. Introduction
The genus Potyvirus, named after its type species, Potato virus Y (PVY) [1], in the family Potyviridae is the largest group of plant RNA viruses that are also economically very important [2]. Potyviruses cause significant loss to agricultural productivity by infecting economically important crops—especially those belonging to the Cucurbitaceae, Solanaceae, Cruciferae and Compositae family [3,4][3][4]. Apart from infecting agriculture crops, potyviruses have wild plant hosts as well [5,6][5][6].
In the Indian subcontinent, pepper/chili is an economically important crop. However, a major bottleneck in its cultivation was crop loss due to infection by viruses, particularly by pepper vein banding virus (PVBV) [3,4][3][4]. PVBV infects Solanaceae plants such as bell-pepper (Capsicum annuum) and chili (Capsicum frutescens) and thus pose a major threat to the agriculture. In bell-pepper, systemic infection of PVBV leads to development of symptoms like mottling, leaf malformation, elongation of petiole and narrowing of leaves [4]. Potyviruses are transmitted by aphids in a non-persistent and non-circulative manner [7] and by seeds to the progeny of infected plants. The aphid transmission takes place by interaction of the virions with aphids via helper component proteinase (HC-Pro) which is a multifunctional potyviral protein having various functional motifs [8]. As with other potyviruses, PVBV virions also bind to aphid mouthparts indirectly through interaction with HC-Pro. The genomic sequence of PVBV was the first potyviral sequence to be reported from India [9,10][9][10]. The 5′-UTR and 3′UTR are 163 and 281 nucleotides in length, respectively, with multiple CAA repeats present in the 5′UTR along with two conserved regions—potybox-a (AACACAACAU) and potybox-b (CUCAAGC) [9]. The 5′ end of the genomic RNA is covalently linked via a phosphodiester linkage to the viral protein genome linked (VPg) and the 3′ end is polyadenylated [9]. Like in other potyviral RNA genomes, PVBV genome encodes for a single ORF which is translated to generate a polyprotein of 3088 amino acids. This polyprotein is processed by the viral-encoded proteases, P1, HC-Pro and NIa-Pro into ten functional and mature proteins [10]. Within the P3 cistron of potyviruses, there is a coding sequence first identified in turnip mosaic virus (TuMV) known as PIPO (pretty interesting potyviridae ORF) [11] [11] which is formed by transcriptional slippage at the conserved G1-2 A6-7 motif in the P3 nucleotide sequence. Another overlapping ORF was identified in the P1 cistron of sweet potato feathery mottle (SPFMV) group of potyvirus, known as PISPO (pretty interesting sweet potato potyvirus ORF) [12[12][13],13], also formed due to the transcriptional slippage at the G1-2 A6-7 slippery sequence in the P1 ORF. Both these ORFs were also located in the PVBV sequence. The genome organization of viruses belonging to Potyvirus genus along with the embedded ORFs and the slippery sequence is shown in Figure 1.
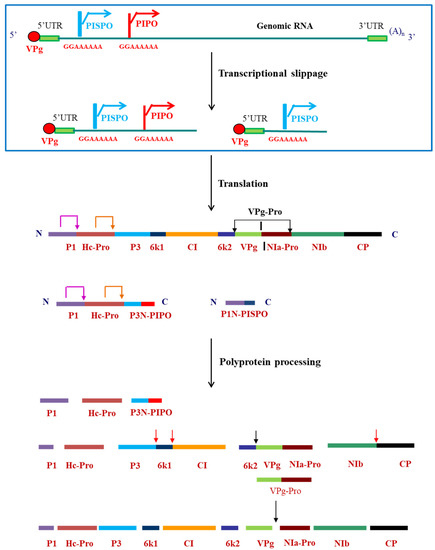
Figure 1. Genome organization of viruses belonging to Potyvirus genus, indicating the transcriptional slippage at the slippery sequences and polyprotein processing by the viral-encoded proteases to form mature proteins. Pink arrow at the P1 and orange arrow at the HC-Pro cleavage sites indicate self-cleavage of the two proteases. Red arrows indicate trans cleavage by VPg-Pro and black arrows indicate the cis cleavage by VPg-Pro.
The proteins encoded by potyviruses are multifunctional, have multiple interacting partner proteins and also participate in various events of the viral life cycle. This functional flexibility could be due to the intrinsically disordered proteins (IDPs)/and intrinsically disordered protein regions (IDPRs) encoded by potyviruses. IDPs are the proteins with large number of distinct and dynamic conformations [14]. This property of intrinsic disorder was specifically studied with PVBV-encoded VPg in the current review. IDPs are natively unfolded proteins which are unfolded along their entire length whereas IDPRs have just a stretch of ≥30 disordered residues within a folded protein [15]. IDPs are involved in one to many and many to one interaction which thus allow the same protein to perform multiple functions. A large proportion of viral proteins exhibit partial or completely disordered structure [16]. The structural disorder in IDPs allow them to interact with their cognate partners with high specificity and low affinity [17]. IDPs undergo a transition to form a more rigid structure known as disorder-to-order transition by virtue of their interaction with specific cognate partner proteins [17,18,19,20][17][18][19][20]. The potyviral proteins such as VPg, CP and P1 have IDPRs which are also known to render multifunctionality to these proteins [21,22,23,24,25,26,27][21][22][23][24][25][26][27]. Both VPg and P1 contain molecular recognition features (MoRFs) in their N-terminal and C-terminal disordered region, respectively [23,28][23][28]. MoRFs are the short regions within the disordered region of a protein which are involved in protein-binding and also undergo a disorder-to-order transition upon binding with a cognate partner protein [29].
2. Applications of Potyviruses in Biotechnology: Characterization of PVBV VLPs as Nanoparticles and Applications of PVBV Chimeric VLPs
Potyviruses and their encoded proteins are used in various biotechnological applications. Different potyviruses are used as vectors for the expression of heterologous proteins in plants. There are insertion sites in potyvirus at the 5′ end of the P1 cistron [84][29], between P1 and HC-Pro [85][30] and between the NIb and CP cistrons [86][31] where various genes can be inserted for their expression. These sites are then proteolytically processed by the viral-encoded proteases, thereby releasing the protein of interest. This property of the potyviruses is therefore exploited for the heterologous expression of proteins. Apart from the virus itself, the potyviral proteins also have a number of other applications. Owing to the high specificity and affinity of NIa-Pro, it is used to remove tags which are added for increasing the solubility and yield of recombinant proteins [87,88,89,90][32][33][34][35]. As mentioned earlier, the CP can self-assemble to form VLPs, therefore these VLPs have been exploited for use as a nanocarrier for a large number of biomedical applications. A large number of plant VLPs/plant virus nanoparticles (PVNs) are used in various biomedical applications such as vaccination, imaging, gene delivery [91][36]. The filamentous rod-shaped viruses have a large surface area to volume ratio, thus providing a high payload capacity and giving them an edge over the icosahedral viruses. There are a few potyviruses which have been studied for various applications in nanotechnology. For instance, PPV and TuMV have been used for antigen presentation and thus a possible application in vaccine [92,93][37][38]. The potyvirus-based nanoparticles have also been used as enzyme nano-carriers (ENCs) where the enzymes of the resveratrol synthetic pathway were immobilized on PVA particles. These immobilized enzymes were thus able to synthesize resveratrol [94][39]. To explore the possibility of PVBV VLPs as nanocarriers, the modes of internalization of PVBV VLPS in different mammalian cells, their intracellular fate and endocytic uptake pathways were studied [95][40]. It was proposed that a clear understanding of nanoparticle-cell interactions is important for developing efficient targeted delivery systems.
Vimentin and Hsp60 were identified as the surface-expressed proteins which facilitated the internalization of PVBV VLPs in HeLa and HepG2 cells, respectively. The major mode of internalization of PVBV VLPs was shown to be caveolae-mediated endocytosis in both HeLa and HepG2 cells. The caveolae-mediated endocytosis is of great importance in nanomedicine since it can bypass the lysosomes, although with few exceptions. Furthermore, this pathway could also be utilized for transvascular delivery of nanomaterials as it is a distinguished transendothelial pathway [96][41]. The animal picornaviruses such as enterovirus 71 and Theiler’s murine encephalomyelitis virus (TMEV) also internalize into infected cells via vimentin [97,98][42][43]. Thus, potyvirus VLPs use the same attachment/entry mechanisms into mammalian cells as animal picornaviruses [99]. [44] It was proposed that PVBV VLPs traffic through endolysosomal pathway after internalization to finally get degraded in the lysosomes. Since PVBV VLPs traffic via endolysosomal pathway, we speculate that this could pave the way for studies on the role of endocytosis in potyvirus infection which remains elusive till date. In the case of TuMV, a plant dynamin-related protein 1 (DRP1, a GTPase involved in endocytosis) from Arabidopsis thaliana (AtDRP1A) was shown to promote TuMV infection. AtDRP1A interacts with VPg and CI, which are both an important component of viral replication complex (VRC). In TuMV-infected cells, it was observed that AtDRP1A also colocalized with VRC. An adaptor protein AP2 was also shown to be an important host factor for TuMV infection [100][45]. Additionally, it was also observed that an inhibition of dynamin not only disrupted endocytosis of TuMV, but also inhibited virus replication and intercellular movement [101][46]. Taken together, these findings thus shed light on the role of components of the endocytic pathway in potyvirus infection.
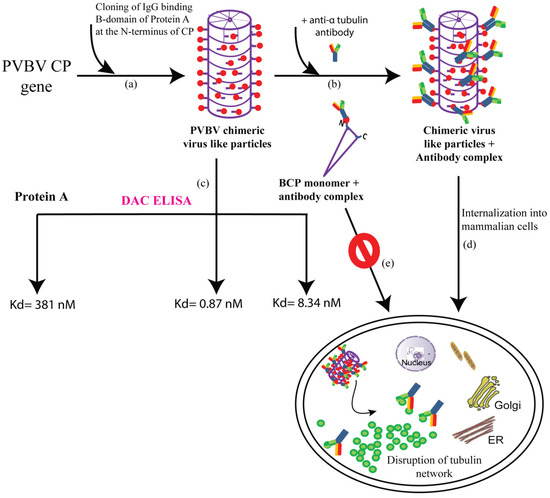
One of the major challenges in immunotherapy, is that the antibodies do not have the ability to enter cells. Therefore, clinical applications of antibodies are limited to those against surface exposed antigens [102][47]. Thus, antibodies to intracellular antigens specific to disease as therapeutics are largely unexplored. With a view to develop PVBV VLP-based nanocarriers for the delivery of antibodies, the B domain of Staphylococcus aureus protein A (SpA) (that binds to the constant region of the antibodies) was genetically engineered at the N-terminus of PVBV CP [103][48]. PVBV CP N-terminal fusion was designated as BCP and assembled virus particles as chimeric PVBV VLPs. The chimeric VLPs generated were shown to have ~500-fold higher IgG-binding affinity than SpA. Such PVBV chimeric VLPs could be of use for developing more sensitive immunodiagnostics. The chimeric PVBV VLPs, like the PVBV VLPs could internalize into various mammalian cells and were degraded after 10 h. The high affinity of the chimeric PVBV VLPs towards antibodies enabled easy generation of chimeric VLP-antibody complexes (for example, anti-CD20 and anti-α tubulin) which have been shown to successfully deliver the bound antibodies intracellularly. Further, the delivered antibodies were also functional inside the cells. Therefore, this property of chimeric PVBV VLPs to bind and deliver antibodies intracellularly could pave the way for future applications in therapeutics [104][49]. Interestingly, only the assembled VLPs could internalize into the cells. BCP purified by Ni-NTA chromatography formed a monomer as per size exclusion chromatography and hence termed as BCP monomer. This BCP monomer failed to interact with vimentin and thus could not internalize into the cells. Thus, it is the assembled scaffold of the viruses that recognize the surface receptors on the host cells and gain entry. The requirement of assembled VLPs also highlights the importance of CP subunits interaction in the development of potyvirus infection. The generation of PVBV chimeric VLPs and their future applications are depicted in Figure 2.
Figure 2. Schematic representation of the generation of chimeric VLPs and their potential applications (a) IgG-binding B-domain is cloned at the N-terminus of CP gene which assembles to form PVBV chimeric VLPs with each subunit expressing the B-domain at its N-terminus (b) Incubating the PVBV chimeric VLPs with anti-α tubulin antibody led to the formation of chimeric VLPs + antibody complex (c) PVBV chimeric VLPs, protein A as well as the monomer BCP (PVBV CP N-terminal fusion with B-domain) subunit + antibody complex were subjected to DAC ELISA (direct antigen coating-enzyme linked immunosorbent assay) where an antibody unrelated to CP was used. The dissociation constant (Kd) for each of the complexes is depicted, indicating an approximately 500-fold higher antibody-binding affinity of chimeric VLPs when compared to protein A (d) chimeric VLPs + anti-α tubulin antibody complex could internalize into mammalian cells and deliver the functional tubulin antibodies, which caused the disruption of the tubulin network ultimately causing cell death (e) whereas the BCP monomer (Ni-NTA purified BCP after size exclusion chromatography) + antibody complex fail to enter the mammalian cells.
3. Conclusions and Future Prospects
Potyvirus biology has been extensively studied and advanced considerably during the last decade. With the increasing evidence of the functions and interacting partners of various potyvirus-encoded proteins, it is now being understood that most of these potyviral proteins are multifunctional playing important roles in more than one viral process. Therefore, the functional regulation of these proteins, by different means such as protein–protein interactions, post-translational modifications, forms an interesting avenue for research that could provide further insights into the complexity of potyviral life cycle.
Understanding the process of transmission of potyviruses, although challenging could open new avenues to control the vector populations as well as dissemination of the potyviral infection in future. Deciphering of viral receptors in the insects is a prerequisite to understand the transmission process. The various animal picornavirus-receptor interactions could be helpful in achieving the same by looking for their counterparts in insects. Internalization of PVBV VLPs via surface-expressed vimentin could also provide some impetus to explore this membrane protein in insects for interaction with potyvirus proteins. While the components of endocytosis during potyviral infection in plant cells is beginning to be identified now, identifying the host proteins that participate in endocytic pathways could also help to control the potyviral infection. As mentioned earlier, AtDRP1A, a host factor involved in TuMV infection could also be a potential target in future for generating potyvirus-resistant plants. Potyviruses exhibit replication-associated translation (RAT) [58][50], however, the coordination between these two steps still remains unanswered. Even the mechanism of switching of VRCs from synthesizing negative-strand viral RNA to the plus-strand RNA needs to be elucidated [104][49].
Although, VPg-Pro has been extensively studied in different potyviruses and is also discussed in this review, still much more remains unexplored. The requirement of ATPase activity of VPg-Pro during different events such as intracellular movement, replication, translation and polyprotein processing, host RNA silencing needs to be understood. This would give mechanistic insights into the potyviral infection. Since ∆N22 VPg has been shown to be a functional globular protein, its crystallization and structure determination can be attempted. Since VPg-Pro has ATPase function, it would also be interesting to examine if VPg-Pro exhibits helicase activity. The motifs and residues involved in the function and regulation of PVBV NIa-Pro and VPg has now been established in a heterologous system. Therefore, it would be interesting to understand if tampering with such motifs/residues would affect PVBV infection.
Flexuous rods are difficult to crystallize, and the cryo EM structure of only two potyviruses, viz watermelon mosaic virus and PVY have been determined so far at 4 Å and 3.4 Å resolution, respectively [105,106][52][53]. Attempts can also be made to crystallize the octameric 16 S ring which is devoid of the N- and C-terminal residues of CP. It would also be interesting to understand how these IDPRs in CP modulate the function of its cognate partner-binding proteins. Both CP and VPg interact with each other and also are involved in the cell-to-cell/long distance movement of the virus, therefore identifying the domain of interaction between them would open new avenues for research in terms of the regulation of the systemic infection of the virus.
The chimeric VLPs of potyviruses hold promise for various biomedical applications. These need to be explored further by in vivo studies.
References
- Ward, C.W.; Shukla, D.D. Taxonomy of potyviruses: current problems and some solutions. Intervirology 1991, 32, 269–96.
- Martínez, F.; Rodrigo, G.; Aragonés, V.; Ruiz, M.; Lodewijk, I.; Fernández, U.; Elena, S.F.; Daròs, J.-A. Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genomics 2016, 17, 87, doi:10.1186/s12864-016-2394-y.
- Revers, F.; García, J.A. Molecular Biology of Potyviruses. Adv. Virus Res. 2015, 92, 101–199, doi:http://dx.doi.org/10.1016/bs.aivir.2014.11.006.
- Ravi, K.S.; Joseph, J.; Nagaraju, N.; Prasad, S.K.; Reddy, H.R.; Savithri, H.S. Characterization of a Pepper Vein Banding Virus from Chili Pepper in India. Plant Dis. 1997, 81, 673–676, doi:10.1094/PDIS.1997.81.6.673.
- Roossinck, M.J. Plant Virus Metagenomics: Biodiversity and Ecology. Annu. Rev. Genet. 2012, 46, 359–369, doi:10.1146/annurev-genet-110711-155600.
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-Wide Variation in Potyviruses. Front. Plant Sci. 2019, 10, 1439, doi:10.3389/fpls.2019.01439.
- King, A.M.Q.; Adams, M.J.; Carsten, E.B.; Lefkowitz, E.J. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses.; 2012; ISBN 9780874216561.
- Valli, A.A.; Gallo, A.; Rodamilans, B.; López-Moya, J.J.; García, J.A. The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763.
- Joseph, J.; Savithri, H.S. Determination of 3′-terminal nucleotide sequence of pepper vein banding virus RNA and expression of its coat protein in Escherichia coli. Arch. Virol. 1999, 144, 1679–1687, doi:10.1007/s007050050696.
- Anindya, R.; Joseph, J.; Gowri, T.D.S.; Savithri, H.S. Complete genomic sequence of Pepper vein banding virus ( PVBV ): a distinct member of the genus Potyvirus. Arch Virol 2004, 149, 625–632, doi:10.1007/s00705-003-0236-0.
- Chung, B.Y.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. 2008, 105, 5897–5902.
- Untiveros, M.; Olspert, A.; Artola, K.; Firth, A.E.; Kreuze, J.F.; Valkonen, J.P.T. A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol. Plant Pathol. 2016, 17, 1111–1123, doi:10.1111/mpp.12366.
- Mingot, A.; Valli, A.; Rodamilans, B.; San León, D.; Baulcombe, D.C.; García, J.A.; López-Moya, J.J. The P1N-PISPO trans-Frame Gene of Sweet Potato Feathery Mottle Potyvirus Is Produced during Virus Infection and Functions as an RNA Silencing Suppressor. J. Virol. 2016, 90, 3543–57, doi:10.1128/JVI.02360-15.
- Fink, A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005, 15, 35–41, doi:10.1016/j.sbi.2005.01.002.
- Uversky, V.N. Intrinsically disordered proteins and their “Mysterious” (meta)physics. Front. Phys. 2019, 7, 10.
- Xue, B.; W. Williams, R.; J. Oldfield, C.; Kian-Meng Goh, G.; Keith Dunker, A.; N. Uversky, V. Viral Disorder or Disordered Viruses: Do Viral Proteins Possess Unique Features? Protein Pept. Lett. 2010, 17, 932–951, doi:10.2174/092986610791498984.
- Uversky, V.N. The Mysterious Unfoldome: Structureless, Underappreciated, Yet Vital Part of Any Given Proteome. J. Biomed. Biotechnol. 2010, 2010, 1–14, doi:10.1155/2010/568068.
- Eliezer, D.; Palmer, A.G. Biophysics: Proteins hunt and gather. Nature 2007, 447, 920–921, doi:10.1038/447920a.
- Uversky, V.N. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, pH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein J. 2009, 28, 305–325, doi:10.1007/s10930-009-9201-4.
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38, doi:10.1016/j.sbi.2008.12.003.
- Makarov, V. V.; Kalinina, N.O. Structure and noncanonical activities of coat proteins of helical plant viruses. Biochem. 2016, 81, 1–18.
- Ksenofontov, A.L.; Paalme, V.; Arutyunyan, A.M.; Semenyuk, P.I.; Fedorova, N. V.; Rumvolt, R.; Baratova, L.A.; Järvekülg, L.; Dobrov, E.N. Partially Disordered Structure in Intravirus Coat Protein of Potyvirus Potato Virus A. PLoS One 2013, 8, e67830, doi:10.1371/journal.pone.0067830.
- Charon, J.; Theil, S.; Nicaise, V.; Michon, T. Protein intrinsic disorder within the Potyvirus genus: from proteome-wide analysis to functional annotation. Mol. Biosyst. 2016, 12, 634–652, doi:10.1039/C5MB00677E.
- Mathur, C.; Jimsheena, V.K.; Banerjee, S.; Makinen, K.; Gowda, L.R.; Savithri, H.S. Functional regulation of PVBV Nuclear Inclusion protein-a protease activity upon interaction with Viral Protein genome-linked and phosphorylation. Virology 2012, 422, 254–264, doi:10.1016/j.virol.2011.10.009.
- Jiang, J.; Laliberté, J.F. The genome-linked protein VPg of plant viruses - A protein with many partners. Curr. Opin. Virol. 2011, 1, 347–354, doi:10.1016/j.coviro.2011.09.010.
- Grzela, R.; Szolajska, E.; Ebel, C.; Madern, D.; Favier, A.; Wojtal, I.; Zagorski, W.; Chroboczek, J. Virulence factor of potato virus Y, genome-attached terminal protein VPg, is a highly disordered protein. J. Biol. Chem. 2008, 283, 213–221, doi:10.1074/jbc.M705666200.
- Rantalainen, K.I.; Christensen, P.A.; Hafrén, A.; Otzen, D.E.; Kalkkinen, N.; Mäkinen, K. Interaction of a potyviral VPg with anionic phospholipid vesicles. Virology 2009, 395, 114–120, doi:10.1016/j.virol.2009.09.009.
- Sabharwal, P.; Srinivas, S.; Savithri, H.S. Mapping the domain of interaction of PVBV VPg with NIa-Pro: Role of N-terminal disordered region of VPg in the modulation of structure and function. Virology 2018, 524, doi:10.1016/j.virol.2018.08.002.
- Katuwawala, A.; Peng, Z.; Yang, J.; Kurgan, L. Computational Prediction of MoRFs, Short Disorder-to-order Transitioning Protein Binding Regions. Comput. Struct. Biotechnol. J. 2019, 17, 454–462.Rajamäki, M.L.; Kelloniemi, J.; Alminaite, A.; Kekarainen, T.; Rabenstein, F.; Valkonen, J.P. A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology 2005, 342, 88–101.
- Klug, A. The tobacco mosaic virus particle: structure and assembly. Phil. Trans. R. Soc. Lond. B 1999, 354, 1–6.Dolja, V.V.; McBride, H.J.; Carrington, J.C. Tagging of plant potyvirus replication and movement by insertion of beta-glucuronidase into the viral polyprotein. Proc. Natl. Acad. Sci. USA 1992, 89, 10208–10212.
- Anindya, R.; Savithri, H.S. Surface-exposed amino- and carboxy-terminal residues are crucial for the initiation of assembly in Pepper vein banding virus : a flexuous rod-shaped virus. Virology 2003, 316, 325–336, doi:10.1016/S0042-6822(03)00593-2.Varrelmann, M.; Palkovics, L.; Maiss, E. Transgenic or plant expression vector-mediated recombination of Plum Pox Virus. J. Virol. 2000, 74, 7462–7469.
- McDonald, M.; Kendall, A.; Bian, W.; McCullough, I.; Lio, E.; Havens, W.M.; Ghabrial, S.A.; Stubbs, G. Architecture of the potyviruses. Virology 2010, 405, 309–313, doi:10.1016/j.virol.2010.06.013.Parks, T.D.; Leuther, K.K.; Howard, E.D.; Johnston, S.A.; Dougherty, W.G. Release of Proteins and Peptides from Fusion Proteins Using a Recombinant Plant Virus Proteinase. Anal. Biochem. 1994, 216, 413–417.
- Ivanov, K.I.; Puustinen, P.; Merits, A.; Saarma, M.; Mäkinen, K. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 2001, 276, 13530–40, doi:10.1074/jbc.M009551200.Kapust, R.B.; Tözsér, J.; Fox, J.D.; Anderson, D.E.; Cherry, S.; Copeland, T.D.; Waugh, D.S. Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001, 14, 993–1000.
- Hervás, M.; Navajas, R.; Chagoyen, M.; García, J.A.; Martínez-Turiño, S. Phosphorylation-related crosstalk between distant regions of the core region of the coat protein contributes to virion assembly of plum pox virus. Mol. Plant-Microbe Interact. 2020, 33, 653–667, doi:10.1094/MPMI-10-19-0305-R.Zheng, N.; de Jesús Pérez, J.; Zhang, Z.; Domínguez, E.; Garcia, J.A.; Xie, Q. Specific and efficient cleavage of fusion proteins by recombinant plum pox virus NIa protease. Protein Expr. Purif. 2008, 57, 153–162.
- Comer, F.I.; Hart, G.W. O-glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J. Biol. Chem. 2000, 275, 29179–29182, doi:10.1074/jbc.R000010200.Nallamsetty, S.; Kapust, R.B.; Tözsér, J.; Cherry, S.; Tropea, J.E.; Copeland, T.D.; Waugh, D.S. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 2004, 38, 108–115.
- Hervás, M.; Ciordia, S.; Navajas, R.; García, J.A.; Martínez‐Turiño, S. Common and Strain‐Specific Post‐Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts. Viruses 2020, 12, doi:10.3390/v12030308.Alemzadeh, E.; Dehshahri, A.; Izadpanah, K.; Ahmadi, F. Plant virus nanoparticles: Novel and robust nanocarriers for drug delivery and imaging. Colloids Surf. B. Biointerfaces 2018, 167, 20–27.
- Pasin, F.; Simón-Mateo, C.; García, J.A. The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses. PLoS Pathog. 2014, 10, e1003985, doi:10.1371/journal.ppat.1003985.Fernández-Fernández, M.R.; Martínez-Torrecuadrada, J.L.; Casal, J.I.; García, J.A. Development of an antigen presentation system based on plum pox potyvirus. FEBS Lett. 1998, 427, 229–235.
- Merits, A.; Lindholm, P.; Runeberg-Roos, P.; Kekarainen, T.; Rajamäki, M.-L.; Puustinen, P.; Mäkeläinen, K.; Saarma, M. Proteolytic processing and membrane-association of potyviral proteins in insect cells. Enviado a Virol. 2000, 1211–1221, doi:10.1099/0022-1317-83-5-1211.Sanchez, F.; Saez, M.; Lunello, P.; Ponz, F. Plant viral elongated nanoparticles modified for log-increases of foreign peptide immunogenicity and specific antibody detection. J. Biotechnol. 2013, 168, 409–415.
- Urcuqui-Inchima, S.; Haenni, A.L.; Bernardi, F. Potyvirus proteins: A wealth of functions. Virus Res. 2001, 74, 157–175, doi:10.1016/S0168-1702(01)00220-9.Besong-Ndika, J.; Wahlsten, M.; Cardinale, D.; Pille, J.; Walter, J.; Michon, T.; Mäkinen, K. Toward the Reconstitution of a Two-Enzyme Cascade for Resveratrol Synthesis on Potyvirus Particles. Front. Plant Sci. 2016, 7, 89.
- Dougherty, W.G.; Parks, T.D. Post-translational processing of the tobacco etch virus 49-kDa small nuclear inclusion polyprotein: identification of an internal cleavage site and delimitation of VPg and proteinase domains. Virology 1991, 183, 449–56.Sabharwal, P.; Amritha, C.K.; Sushmitha, C.; Natraj, U.; Savithri, H.S. Intracellular trafficking and endocytic uptake pathway of Pepper vein banding virus-like particles in epithelial cells. Nanomedicine 2019, 14, 1247–1265.
- Joseph, J.; Savithri, H.S. Mutational analysis of the NIa protease from pepper vein banding potyvirus. Arch. Virol. 2000, 145, 2493–2502, doi:10.1007/s007050070004.Sahay, G.; Alakhova, D.Y.; Kabanov, A.V. Endocytosis of Nanomedicines. J. Control. Release 2010, 145, 182–195.
- Kim, D.; Kang, B.H.; Han, J.S.; Choi, K.Y. Temperature and salt effects on proteolytic function of turnip mosaic potyvirus nuclear inclusion protein a exhibiting a low-temperature optimum activity. Biochim. Biophys. Acta 2000, 1480, 29–40.Du, N.; Cong, H.; Tian, H.; Zhang, H.; Zhang, W.; Song, L.; Tien, P. Cell Surface Vimentin Is an Attachment Receptor for Enterovirus 71. J. Virol. 2014, 88, 5816–5833.
- Rorrer, K.; Parks, T.D.; Scheffler, B.; Bevan, M.; Dougherty, W.G. Autocatalytic activity of the tobacco etch virus NIa proteinase in viral and foreign protein sequences. J. Gen. Virol. 1992, 73, 775–783.Nédellec, P.; Vicart, P.; Laurent-Winter, C.; Martinat, C.; Prévost, M.C.; Brahic, M. Interaction of Theiler’s virus with intermediate filaments of infected cells. J. Virol. 1998, 72, 9553–9560.
- Schaad, M.C.; Haldeman-Cahill, R.; Cronin, S.; Carrington, J.C. Analysis of the VPg-proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 1996, 70, 7039–48.Koudelka, K.J.; Destito, G.; Plummer, E.M.; Trauger, S.A.; Siuzdak, G.; Manchester, M. Endothelial Targeting of Cowpea Mosaic Virus (CPMV) via Surface Vimentin. PLoS Pathog. 2009, 5, e1000417.
- Carrington, J.C.; Haldeman, R.; Dolja, V. V; Restrepo-Hartwig, M.A. Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J. Virol. 1993, 67, 6995–7000, doi:10.1128/jvi.67.12.6995-7000.1993.Wu, G.; Cui, X.; Dai, Z.; He, R.; Li, Y.; Yu, K.; Bernards, M.; Chen, X.; Wang, A. A plant RNA virus hijacks endocytic proteins to establish its infection in plants. Plant J. 2019, 101, 384–400.
- Carrington, J.C.; Cary, S.M.; Parks, T.D.; Dougherty, W.G. A second proteinase encoded by a plant potyvirus genome. EMBO J. 1989, 8, 365–370.Wu, G.; Cui, X.; Chen, H.; Renaud, J.B.; Yu, K.; Chen, X.; Wang, A. Dynamin-Like Proteins of Endocytosis in Plants Are Coopted by Potyviruses To Enhance Virus Infection. J. Virol. 2018, 92.
- Ghabrial, S.A.; Smith, H.A.; Parks, T.D.; Dougherty, W.G. Molecular genetic analyses of the soybean mosaic virus NIa protease. J. Gen. Virol. 1990, 71, 1921–1927, doi:10.1099/0022-1317-71-9-1921.Slastnikova, T.A.; Ulasov, A.V.; Rosenkranz, A.A.; Sobolev, A.S. Targeted Intracellular Delivery of Antibodies: The State of the Art. Front. Pharmacol. 2018, 9, 1208.
- Dougherty, W.G.; Carrington, J.C. Expression and Function of Potyviral Gene Products. Annu. Rev. Phytopathol. 1988, 26, 123–143, doi:10.1146/annurev.py.26.090188.001011.Sabharwal, P.; Sushmitha, C.; Amritha, C.K.; Natraj, U.; Murthy, M.R.N.; Savithri, H.S. Development of pepper vein banding virus chimeric virus-like particles for potential diagnostic and therapeutic applications. Arch. Virol. 2020, 165, 1163–1176.
- Phan, J.; Zdanov, A.; Evdokimov, A.G.; Tropea, J.E.; Iii, H.K.P.; Kapust, R.B.; Li, M.; Wlodawer, A.; Waugh, D.S. Structural Basis for the Substrate Specificity of Tobacco Etch Virus Protease . J. Biol. Chem. 2002, 277, 50564–50572, doi:10.1074/jbc.M207224200.Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-dependent RNA polymerase NiB of potyviruses plays multifunctional, contrasting roles during viral infection. Viruses 2020, 12, 77.
- Sun, P.; Austin, B.P.; Tözsér, J.; Waugh, D.S. Structural determinants of tobacco vein mottling virus protease substrate specificity. Protein Sci. 2010, 19, 2240–2251, doi:10.1002/pro.506.Léonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberté, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7737.
- Chouard, T. Structural biology: Breaking the protein rules. Nature 2011, 471, 151–153, doi:10.1038/471151a.Zamora, M.; Méndez-López, E.; Agirrezabala, X.; Cuesta, R.; Lavín, J.L.; Sánchez-Pina, M.A.; Aranda, M.A.; Valle, M. Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv. 2017, 3, eaao2182.
- Mathur, C.; Savithri, H.S. Novel ATPase activity of the polyprotein intermediate, Viral Protein genome-linked-Nuclear Inclusion-a protease, of Pepper vein banding potyvirus. Biochem. Biophys. Res. Commun. 2012, 427, 113–118, doi:10.1016/j.bbrc.2012.09.020.Kežar, A.; Kavčič, L.; Polák, M.; Nováček, J.; Gutiérrez-Aguirre, I.; Žnidarič, M.T.; Coll, A.; Stare, K.; Gruden, K.; Ravnikar, M.; et al. Structural basis for the multitasking nature of the potato virus Y coat protein. Sci. Adv. 2019, 5, eaaw3808.
- Eskelin, K.; Hafren, A.; Rantalainen, K.I.; Makinen, K. Potyviral VPg Enhances Viral RNA Translation and Inhibits Reporter mRNA Translation In Planta. J. Virol. 2011, 85, 9210–9221, doi:10.1128/JVI.00052-11.Kežar, A.; Kavčič, L.; Polák, M.; Nováček, J.; Gutiérrez-Aguirre, I.; Žnidarič, M.T.; Coll, A.; Stare, K.; Gruden, K.; Ravnikar, M.; et al. Structural basis for the multitasking nature of the potato virus Y coat protein. Sci. Adv. 2019, 5, eaaw3808.
- Rajamäki, M.-L.; Valkonen, J.P.T. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 2009, 21, 2485–502, doi:10.1105/tpc.108.064147.
- Rantalainen, K.I.; Eskelin, K.; Tompa, P.; Mäkinen, K. Structural flexibility allows the functional diversity of potyvirus genome-linked protein VPg. J. Virol. 2011, 85, 2449–57, doi:10.1128/JVI.02051-10.
- Dunoyer, P.; Thomas, C.; Harrison, S.; Revers, F.; Maule, A. A Cysteine-Rich Plant Protein Potentiates Potyvirus Movement through an Interaction with the Virus Genome-Linked Protein VPg. J. Virol. 2004, 78, 2301–2309, doi:10.1128/JVI.78.5.2301.
- Grzela, R.; Strokovska, L.; Andrieu, J.P.; Dublet, B.; Zagorski, W.; Chroboczek, J. Potyvirus terminal protein VPg, effector of host eukaryotic initiation factor eIF4E. Biochimie 2006, 88, 887–896, doi:10.1016/j.biochi.2006.02.012.
- Léonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberté, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7, doi:10.1128/JVI.74.17.7730-7737.2000.
- Hébrard, E.; Bessin, Y.; Michon, T.; Longhi, S.; Uversky, V.N.; Delalande, F.; Van Dorsselaer, A.; Romero, P.; Walter, J.; Declerck, N.; et al. Intrinsic disorder in Viral Proteins Genome-Linked: experimental and predictive analyses. Virol. J. 2009, 6, 23, doi:10.1186/1743-422X-6-23.
- Satheshkumar, P.S.; Gayathri, P.; Prasad, K.; Savithri, H.S. “Natively unfolded” VPg is essential for Sesbania mosaic virus serine protease activity. J. Biol. Chem. 2005, 280, 30291–30300, doi:10.1074/jbc.M504122200.
- Schein, C.H.; Oezguen, N.; Volk, D.E.; Garimella, R.; Paul, A.; Braun, W. NMR structure of the viral peptide linked to the genome (VPg) of poliovirus. Peptides 2006, doi:10.1016/j.peptides.2006.01.018.
- Fernández, A.; Guo, H.S.; Sáenz, P.; Simón-buela, L.; Cedrón, M.G. De; García, J.A. The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. 1997, 25, 4474–4480.
- Guo, D.; Rajamäki, M.L.; Saarma, M.; Valkonen, J.P.T. Towards a protein interaction map of potyviruses: Protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 2001, 82, 935–939, doi:10.1099/0022-1317-82-4-935.
- Klein, P.G.; Klein, R.R.; Rodríguez-Cerezo, E.; Hunt, A.G.; S.J. Mutational Analysis of the Tobacco Vein Mottling Virus Genome. Virology 1994, 204, 759–769, doi:10.1006/VIRO.1994.1591.
- Riechmann, J.L.; Lain, S.; Garcia, J.A. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 1992, 73, 1–16, doi:10.1099/0022-1317-73-1-1.
- Hong Y, H.A. RNA Polymerase Activity Catalyzed by a Potyvirus-Encoded RNA-Dependent RNA Polymerase. Virology 1996, 226, 146–151, doi:10.1006/VIRO.1996.0639.
- Anindya, R.; Chittori, S.; Savithri, H.S. Tyrosine 66 of Pepper vein banding virus genome-linked protein is uridylylated by RNA-dependent RNA polymerase. Virology 2005, 336, 154–162, doi:10.1016/j.virol.2005.03.024.
- Puustinen, P.; Mäkinen, K. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 2004, 279, 38103–38110, doi:10.1074/jbc.M402910200.
- Carette, J.E.; Kujawa, A.; Gühl, K.; Verver, J.; Wellink, J.; V.K.A. Mutational Analysis of the Genome-Linked Protein of Cowpea Mosaic Virus. Virology 2001, 290, 21–29, doi:10.1006/VIRO.2001.1137.
- Murray, K.E.; Barton, D.J. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 2003, 77, 4739–50, doi:10.1128/JVI.77.8.4739-4750.2003.
- Paul, A. V.; van Boom, J.H.; Filippov, D.; Wimmer, E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 1998, 393, 280–284, doi:10.1038/30529.
- Cheng, X.; Xiong, R.; Li, Y.; Li, F.; Zhou, X.; Wang, A. Sumoylation of turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 2017, 29, 508–525, doi:10.1105/tpc.16.00774.
- Li, F.; Zhang, C.; Li, Y.; Wu, G.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1–17, doi:10.1038/s41467-018-03658-2.
- Cheng, X.; Wang, A. The Potyvirus Silencing Suppressor Protein VPg Mediates Degradation of SGS3 via Ubiquitination and Autophagy Pathways. J. Virol. 2017, 91, doi:10.1128/jvi.01478-16.
- Kushwaha, N.K.; Hafrén, A.; Hofius, D. Autophagy-virus interplay in plants: from antiviral recognition to proviral manipulation. Mol. Plant Pathol. 2019, 20, 1211–1216, doi:10.1111/mpp.12852.
- Langenberg, W.G.; Zhang, L. Immunocytology shows the presence of tobacco etch virus P3 protein in nuclear inclusions. J. Struct. Biol. 1997, 118, 243–7, doi:10.1006/jsbi.1997.3856.
- Anindya, R.; Savithri, H.S. Potyviral NIa Proteinase, a Proteinase with Novel Deoxyribonuclease Activity. J. Biol. Chem. 2004, 279, 32159–32169, doi:10.1074/jbc.M404135200.
- Riechmann, J.L.; Cervera, M.T.; Garcia, J.A. Processing of the plum pox virus polyprotein at the P3-6K1 junction is not required for virus viability. J. Gen. Virol. 1995, 76, 951–956, doi:10.1099/0022-1317-76-4-951.
- Hafren, A.; Hofius, D.; Ronnholm, G.; Sonnewald, U.; Makinen, K. HSP70 and Its Cochaperone CPIP Promote Potyvirus Infection in Nicotiana benthamiana by Regulating Viral Coat Protein Functions. Plant Cell 2010, 22, 523–535, doi:10.1105/tpc.109.072413.
- Besong-Ndika, J.; Ivanov, K.I.; Hafrèn, A.; Michon, T.; Mäkinen, K. Cotranslational Coat Protein-Mediated Inhibition of Potyviral RNA Translation. J. Virol. 2015, 89, 4237–4248, doi:10.1128/JVI.02915-14.
- Puustinen, P.; Rajamäki, M.-L.; Ivanov, K.I.; Valkonen, J.P.T.; Mäkinen, K. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J. Virol. 2002, 76, 12703–11.
- Ivanov, K.I.; Puustinen, P.; Gabrenaite, R.; Vihinen, H.; Rönnstrand, L.; Valmu, L.; Kalkkinen, N.; Mäkinen, K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell 2003, 15, 2124–39.
- Xiong, R.; Wang, A. SCE1, the SUMO-Conjugating Enzyme in Plants That Interacts with NIb, the RNA-Dependent RNA Polymerase of Turnip Mosaic Virus, Is Required for Viral Infection. J. Virol. 2013, 87, 4704–4715, doi:10.1128/JVI.02828-12.
- Rajamäki, M.L.; Kelloniemi, J.; Alminaite, A.; Kekarainen, T.; Rabenstein, F.; V.J. A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology 2005, 342, 88–101, doi:10.1016/J.VIROL.2005.07.019.
- Dolja, V. V; McBride, H.J.; Carrington, J.C. Tagging of plant potyvirus replication and movement by insertion of beta-glucuronidase into the viral polyprotein. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 10208–12.
- Varrelmann, M.; Palkovics, L.; Maiss, E. Transgenic or plant expression vector-mediated recombination of Plum Pox Virus. J. Virol. 2000, 74, 7462–9.
- Parks, T.D.; Leuther, K.K.; Howard, E.D.; Johnston, S.A.; Dougherty, W.G. Release of Proteins and Peptides from Fusion Proteins Using a Recombinant Plant Virus Proteinase. Anal. Biochem. 1994, 216, 413–417, doi:10.1006/abio.1994.1060.
- Kapust, R.B.; Tözsér, J.; Fox, J.D.; Anderson, D.E.; Cherry, S.; Copeland, T.D.; Waugh, D.S. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001, 14, 993–1000.
- Zheng, N.; Pérez, J. de J.; Zhang, Z.; Domínguez, E.; Garcia, J.A.; Xie, Q. Specific and efficient cleavage of fusion proteins by recombinant plum pox virus NIa protease. Protein Expr. Purif. 2008, 57, 153–162, doi:10.1016/j.pep.2007.10.008.
- Nallamsetty, S.; Kapust, R.B.; Tözsér, J.; Cherry, S.; Tropea, J.E.; Copeland, T.D.; Waugh, D.S. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 2004, 38, 108–115, doi:10.1016/j.pep.2004.08.016.
- Alemzadeh, E.; Dehshahri, A.; Izadpanah, K.; Ahmadi, F. Plant virus nanoparticles: Novel and robust nanocarriers for drug delivery and imaging. Colloids Surf. B. Biointerfaces 2018, 167, 20–27, doi:10.1016/j.colsurfb.2018.03.026.
- Fernández-Fernández, M.R.; Martínez-Torrecuadrada, J.L.; Casal, J.I.; García, J.A. Development of an antigen presentation system based on plum pox potyvirus. FEBS Lett. 1998, 427, 229–35.
- Sanchez, F.; Saez, M.; Lunello, P.; Ponz, F. Plant viral elongated nanoparticles modified for log-increases of foreign peptide immunogenicity and specific antibody detection. J. Biotechnol. 2013, 168, 409–415, doi:10.1016/j.jbiotec.2013.09.002.
- Besong-Ndika, J.; Wahlsten, M.; Cardinale, D.; Pille, J.; Walter, J.; Michon, T.; Mäkinen, K. Toward the Reconstitution of a Two-Enzyme Cascade for Resveratrol Synthesis on Potyvirus Particles. Front. Plant Sci. 2016, 7, 89, doi:10.3389/fpls.2016.00089.
- Sabharwal, P.; Amritha, C.K.; Sushmitha, C.; Natraj, U.; Savithri, H.S. Intracellular trafficking and endocytic uptake pathway of Pepper vein banding virus-like particles in epithelial cells. Nanomedicine (Lond). 2019, 14, 1247–1265, doi:10.2217/nnm-2018-0405.
- Sahay, G.; Alakhova, D.Y.; Kabanov, A. V Endocytosis of Nanomedicines. 2010, doi:10.1016/j.jconrel.2010.01.036.
- Du, N.; Cong, H.; Tian, H.; Zhang, H.; Zhang, W.; Song, L.; Tien, P. Cell Surface Vimentin Is an Attachment Receptor for Enterovirus 71. J. Virol. 2014, 88, 5816–5833, doi:10.1128/JVI.03826-13.
- Nédellec, P.; Vicart, P.; Laurent-Winter, C.; Martinat, C.; Prévost, M.C.; Brahic, M. Interaction of Theiler’s virus with intermediate filaments of infected cells. J. Virol. 1998, 72, 9553–60.
- Koudelka, K.J.; Destito, G.; Plummer, E.M.; Trauger, S.A.; Siuzdak, G.; Manchester, M. Endothelial Targeting of Cowpea Mosaic Virus (CPMV) via Surface Vimentin. PLoS Pathog. 2009, 5, e1000417, doi:10.1371/journal.ppat.1000417.
- Wu, G.; Cui, X.; Dai, Z.; He, R.; Li, Y.; Yu, K.; Bernards, M.; Chen, X.; Wang, A. A plant RNA virus hijacks endocytic proteins to establish its infection in plants. Plant J. 2019, 101, 384–400, doi:10.1111/tpj.14549.
- Wu, G.; Cui, X.; Chen, H.; Renaud, J.B.; Yu, K.; Chen, X.; Wang, A. Dynamin-Like Proteins of Endocytosis in Plants Are Coopted by Potyviruses To Enhance Virus Infection. J. Virol. 2018, 92, doi:10.1128/jvi.01320-18.
- Slastnikova, T.A.; Ulasov, A. V; Rosenkranz, A.A.; Sobolev, A.S. Targeted Intracellular Delivery of Antibodies : The State of the Art. Front. Pharmacol. 2018, 9, 1–21, doi:10.3389/fphar.2018.01208.
- Sabharwal, P.; Sushmitha, C.; Amritha, C.K.; Natraj, U.; Murthy, M.R.N.; Savithri, H.S. Development of pepper vein banding virus chimeric virus-like particles for potential diagnostic and therapeutic applications. Arch. Virol. 2020, doi:10.1007/s00705-020-04581-y.
- Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-dependent RNA polymerase NiB of potyviruses plays multifunctional, contrasting roles during viral infection. Viruses 2020, 12, doi:10.3390/v12010077.
- Zamora, M.; Méndez-López, E.; Agirrezabala, X.; Cuesta, R.; Lavín, J.L.; Sánchez-Pina, M.A.; Aranda, M.A.; Valle, M. Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv. 2017, 3, eaao2182, doi:10.1126/sciadv.aao2182.