Today, researchers are looking at new ways to treat severe infections caused by resistance to standard antibiotic therapy. This is quite challenging due to the complex and interdependent relationships involved: the cause of infection–the patient–antimicrobial agents. The sessile biofilm form is essential in research to reduce resistance to very severe infections (such as ESKAPE pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanni, Pseudomonas aeruginosa, and Enterobacter spp). The purpose of this study is to elucidate the mechanisms of the occurrence, maintenance, and suppression of biofilm infections. One form of biofilm suppression is the efficient action of natural antagonists of bacteria—bacteriophages. Bacteriophages effectively penetrate the biofilm’s causative cells. They infect those bacterial cells and either destroy them or prevent the infection from spreading. In this process, bacteriophages are specific, relatively easy to apply, and harmless to the patient. Antimicrobial peptides (AMPs) support the mechanisms of bacteriophages’ action. AMPs could also attack and destroy infectious agents on their own (even on biofilm). AMPs are simple, universal peptide molecules, mainly cationic peptides. Additional AMP research could help develop even more effective treatments of biofilm (bacteriophages, antibiotics, AMPs, nanoparticles). This is a review of recent unconventional agents, such as bacteriophages and AMPs, used for the eradication of biofilm, providing an overview of potentially new biofilm treatment strategies.
- antimicrobial peptides
- bacteriophage
- biofilm
- immunomodulatory action
- resistance
1. Introduction
From its beginning until the present day, the natural course of human life has been marked by transition, and it is still passing through often unknown processes of adaptation and evolution [1]. Destroying life, and even destroying the smallest carriers—microorganisms, is not easy and very often impossible [2]. Is it possible at all, and for how long, to delay an event's unfavorable and unwanted course? The ineffectiveness of antibacterial drugs is not an isolated phenomenon but an increasingly common occurrence [3,4,5][3][4][5]. Increasing bacterial resistance is connected to the patient and clinicians’ malpractice in prescribing and using antibiotics [6].
When science created the first antibiotics preparations to facilitate and raise human life quality, humanity was relieved [7]. Deadly diseases became transient conditions, and a growing selection of effective drugs guaranteed an optimistic future and extended life expectancy [8]. Simultaneously, the ever deeper delving began into the unknown principles of maintenance of life as a phenomenon.
Slowly, the growing lack of antibiotic effectiveness has led us along the path of learning about the mechanism of adaptation and even the evolution of the bacteria that carry the simplest forms of life, returning us to the very beginning and even simpler forms of life—viruses. Thus, the appearance of resistance in bacteria demonstrates one of the fundamental principles of preserving the phenomenon of life [9]. Thanks to technological advances, new ways of delivering antimicrobial peptides have been developed—one is by using nanoparticles. Those with silver are the best choice due to its antimicrobial activity [10,11,12,13][10][11][12][13].
2. Biofilm Treatment Strategies
The ability to make biofilm is an evolutionary achievement, with new “multicellular ability” traits that allow bacteria to survive, infect, multiply, and permanently infect hosts. As already mentioned, the resistance of clinically relevant bacteria (ESKAPE) and other groups of resistant bacteria is primarily the ability to infect the host despite biofilm inhibition measures taken, such as surface change and modification (medical implants or other biomaterials) using antibacterial agents, where the coating creates a barrier to bacterial adhesion [14][15][16]. Also, the use of small molecules of bacterial biofilm inhibitors creates antifilm properties that passivate the surface of implants or medical devices (such as phenols, imidazoles, indole) [17][18][19]. An alternative method in biofilm control is biologically active agents, such as predatory bacteria species [20].
Biofilm dispersion is the second strategy in treating biofilm infections. More precisely, the disruption of quorum sensing by chemical means leads to biofilm dispersion [21][22][23]. However, biofilm dispersal agents should be combined with an antimicrobial agent [17][24]. Namely, if these dispersed bacteria are not treated simultaneously with antibacterial agents, they will inevitably form new biofilms by infecting new areas [25][16]. Treatment by co-administration of drugs and dispersal agents is very complex and challenging. However, as usual, the answer to this phenomenon already exists in microbiocenosis, by “infecting” the biofilm with viruses—bacteriophages.
2.1. Bacteriophages
2.1. Bacteriophages
Bacteriophages are viruses that infect bacteria. Bacteriophages have been infectiously parasitic on bacteria from the very beginning of life. This virus–bacterium relationship is the oldest form of microbiocenosis, and perfection has been achieved in the form of a specific match between the virus –bacteriophage and the host cell-bacterium [26][27][28].
The first practical and positive experiences of using bacteriophages in controlling bacterial infections were developed during the Second World War. Bacteriophages found their application in treating war wound infections (explosive and blast injuries) before the use of antibiotics [29]. In extensive infections of such wounds, pathogen-specific bacteriophage preparations (
,
,
,
spp.) were applied directly to the site of infection (biofilm infections) [30].
The phage action mechanism on the prokaryotic cell begins with overcoming the cell membrane’s defense mechanisms, the incorporation of the genome into the cytoplasm, and the proliferation of phages. Phages impair the bacterial cells’ normal function by their proliferation and thus inactivate or kill cells (lysogenic or lytic cycle,
) [26][29]. The relationship between bacteriophages and bacteria in biofilm is far more complex. Bacteriophages must have the ability to encode a depolymerase that degrades the biofilm matrix, which includes degradation of polymers, capsular polysaccharides, and extracellular DNA [31][32]. Only then do they access cell membranes and receptors. In the treatment of bacterial infections, the addition of several enzymes that increases phages' activity is used, which leads to synergistic removal of bacteria. Thus, it is essential to create the conditions to bypass the bacterial biofilm matrix [33][34]. Therefore, bacteriophages can penetrate the membrane receptors, but antibiotics cannot due to the biofilm’s defense mechanisms [35][36]. The combination of phage and antibiotics seems to be the optimal combination in the fight against biofilm. In most cases, it is optimal in various combinations, even in combination with disinfectants [37][38]. However, in some situations, phage application could even lead to enhanced bacteria aggregation in the biofilm, surface adhesion, and fimbriae production. This usually occurs in Gram-negative bacteria leading to inhibition of phage penetration through the biofilm [33][39][40].
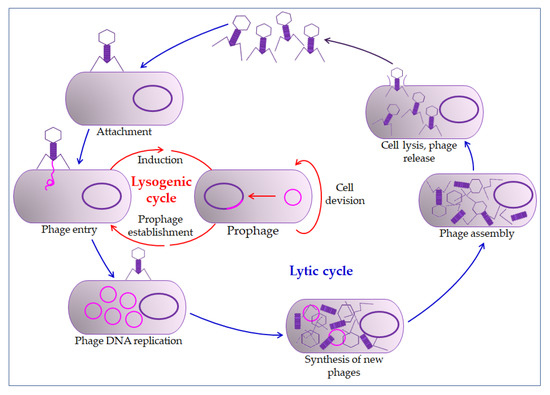
Bacteriophage life cycle. The bacteriophage first interacts with receptors on the host, absorbs, and then injects its genome to infect a bacterium. The lytic cycle involves the production of new bacteriophages and their release from the infected cell by lysis. The lysogenic cycle results in integrating a phage genome into the bacterial genome, which replicates in concert with the host DNA.
Furthermore, there are circumstances in which phages stimulate biofilm formation, such as increased bacteriophage pressure in the biofilm, leading to larger aggregates, which could be considered an evolutionary adaptation [33][39]. Thus, such interaction between bacteriophage and the host could be classified as mutualistic rather than parasitic. In so doing, bacteriophages acquire some new properties which are favorable to them, such as encapsulation in the biofilm matrix, in which phages can tolerate higher concentrations of disinfectants, radiation, and other environmental factors [35][33].
All this points to the need for careful preparation before the application of bacteriophages. Due to prophage induction, extracellular DNA accumulates in the biofilm [33][41]. Although they could lead to cell death, prophages are integrated phages in the host’s lysogenic cycle. For example, they may encode virulence factors and antibiotic resistance factors (toxins, enzymes, and superantigens) in cholera [33].
Bacteriophage evolution stimulated by phage and host interaction in the biofilm potentiates and promotes mutations as common properties. It could demonstrate the real potential of bacteriophage therapy to eradicate infectious biofilms [38][42]. Thus, hundreds of years of positive practical experience in applying bacteriophages, their easy isolation, cost-effectiveness, absolute specificity to the host, self-reproduction, and non-disruption of normal microbiocenosis, without harmful side effects, make bacteriophages the choice for the future [26][43].
There are a few more potential benefits of bacteriophages. The ability to “deliver” broad-spectrum antimicrobial drugs to the infection site makes bacteriophages extremely potent in creating even more effective modular antibacterial agents [44]. Another particular interest is the concept of enhancing the phage genome to express antimicrobial peptides (AMPs) [14].
2.2. Antimicrobial Peptides (AMPs)
2.2. Antimicrobial Peptides (AMPs)
Biofilm eradication agents (BEAs) are the target of many modern studies, and antimicrobial peptides (AMPs) are among the most likely BEAs [17][45][46][47]. AMPs are ubiquitous compounds produced by plants, invertebrates, and animals [48] and are relatively simple molecules (from 5 to 100 amino acids) with a molecular mass of 1–5 kDa [14][49]. They are predominantly cationic, so they are also called cationic antimicrobial peptides. Their antimicrobial action mechanism is associated mainly with cytoplasmic membrane disorder and inhibition of protein accumulation or enzymatic activity. In eukaryotes, they play an essential role in innate immunity [50][51].
Plants and invertebrates lack adaptive immunity (immunity mediated by B and T cells), so AMPs play a fundamental role in protecting against bacterial and fungal infections [48]. All plant AMPs are rich in cysteine and contain many disulfide bonds. In invertebrates, AMPs are found in hemolymph, hemocytes, phagocytes, and epithelial cells [48][49]. The vertebrate immune system consists of an innate and adaptive immune system. AMPs can be isolated from leukocytes, phagocytes, epithelial macrophages, and body fluids [49][52][53][54]. The most prominent groups of mammalian AMPs are cathelicidins and defensins [50][55].
AMPs are classified according to structure, sequence, or mechanism of action. AMPs may have several activities: bactericidal, immune modulations, antiviral properties, anticarcinogenic properties, and they can prevent biofilm formation. Since AMP activity depends on their structure and sequence, it is crucial to consider both properties when categorizing them [48][49].
2.2.1. The Mechanism of Action of Antimicrobial Peptides
The mechanism of action of AMPs can be divided into two groups: the direct killing of microorganisms (by membrane targeting or non-membrane targeting) or immune modulation [14][51][56].
The permeabilizing membrane mechanism of action may have receptor- or non-receptor-mediated interactions. Some AMPs, such as nisin, bind with high affinity to lipid molecules in the cell membrane, producing pores in it, and act by covering the entire surface of the membrane, the so-called carpet model (detergent-like model) [22][48][57].
The direct non-membrane targeting mechanisms of action are based on AMPs targeting the bacterial cell wall to inhibit cell wall synthesis. AMPs interact with the diverse precursors needed for cell wall synthesis. For example, AMP defensins bind to the charged pyrophosphate sugar moiety of the lipid molecule [48][58]. While some AMPs can act on the cytoplasmic membrane, others accumulate in the cytoplasm and inhibit proteins and nucleic acid synthesis, thus disrupting enzyme–protein activity (
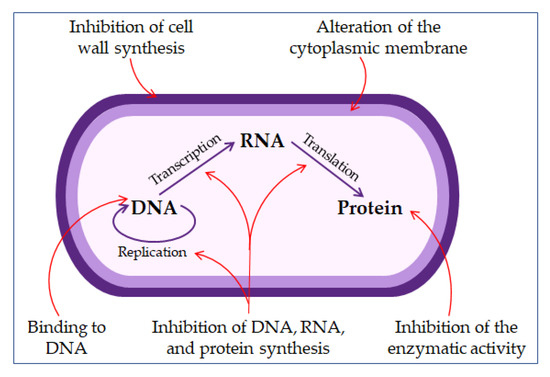
The mechanisms of antimicrobial peptides action.
The mechanisms of action of AMP in immune modulation include various immune responses. Immune cells (neutrophils, macrophages) produce AMPs, which are the first to contact the microbial invasion [60][61]. Likewise, AMPs promote a diversity of immune responses: activation, attraction, and differentiation of leukocytes. Some human AMPs (LL-37, β defensins) can attract immune cells, such as white blood cells, dendritic cells, and mast cells [48][62][63][64]. Some research suggests that AMPs might serve as vaccine adjuvants [
].
The properties of AMPs can be improved to enhance their delivery by optimizing their stability and toxicity. This is mainly done through polymer conjugation: conjugation with biopolymers such as chitosan and hyaluronic acid. Alternatively, it may be done by encapsulating AMPs in micelles and liposomes [48][65].
Antiviral AMPs can neutralize the virus by integrating into the viral envelope and cell membrane, causing the viral membrane's destabilization or preventing the host’s infection [62]. Antiviral AMPs defensins can bind to viral glycoprotein after which viruses (such as herpes simplex virus) cannot bind to the surface of the host cell [55]. Some AMPs (such as lactoferrin) can occupy specific mammalian cell receptors and prevent the virus from binding to its target receptor (such as heparan molecules for herpes simplex virus) and blocking viral interaction with the receptor [47][49]. Some antiviral AMPs can enter the host cell itself where they are located in the cytoplasm or the organelles and alter the host cell gene expression, thus helping the host defense mechanism [47][66][67].
Thus, AMPs, with all their properties and mechanisms of action (structural, therapeutic), are incredibly suitable molecules in the treatment, especially of infections resistant to many drugs (mainly resistant to antibiotics) [68][69][70][71]. All future research should aim at discovering the improvement of AMPs' intake and their action, and their action combined with other antimicrobial agents (antibiotics, bacteriophages) [72][73][74]. This mainly refers to their biocompatibility action in the immunomodulation system [46][64][70]. It is also necessary to avoid undesirable consequences of AMP administration such as toxicity, hemolytic activity, and changing their structure, primarily of cationic AMPs, to obtain even more efficient and safer AMPs [71].
2.2.2. The Benefit of Combined Therapy of Antimicrobial Peptides and Nanoparticles
New AMP delivery systems are being developed, which could help avoid the problems related to AMP delivery [14][75]. They improve the pharmacokinetics of AMPs [10], increase their half-life, reduce the required dose, and decrease production costs and possible toxicity [10][11][76]. All this may be achieved by encapsulating AMPs in various nanocarriers [14][75]. Nanoparticles significantly increase the penetration of AMPs into cells [77][76].
Several metal nanoparticles, such as silver and gold, have appeared as a possible choice for treating antibiotic-resistant bacterial infections [12]. Silver nanoparticles (AgNPs) are particularly interesting because they have potent antimicrobial activity [12][78]. Both AgNPs and AMPs could replace antibiotics, and the conjugation of AMPs with AgNPs has the additional advantage of the synergistic effect of their antimicrobial properties [79][80][81]. The combination of AMPs and AgNPs might produce new features, such as higher antibacterial activity, increased stability, reduced toxicity, and enhanced selectivity [13][82][83].
Combined therapy using AgNPs and AMPs represents a new approach to developing new antimicrobial drugs [84].
3. Conclusions
Today’s findings on microbial diseases (primarily bacterial) indicate the constant dynamic of microorganisms in adaptation to antimicrobial drugs. The most harmful and clinically significant pathogens are classified into resistant groups (ESKAPE, MRSA, VRE) and create a biofilm as a biological response to drugs. They acquire various forms of resistance by rapid mutations, changes in antigen structure, and adapt their virulence mechanisms and contagiousness. By studying these models, mechanisms, and principles, treatment options arise from the microorganisms’ environment. One is the use of the evolutionary abilities of some other microorganisms. Thus, bacterial antagonists, bacteriophages, and their infection mechanisms and parasitism on bacteria are used to improve the treatment of severe infections.
This principle that nature offers solutions exactly where problems arise is combined with increasing knowledge about relatively simple proteins. AMPs can act alone or in combination with known or innovated antibiotics, bacteriophages, and nanoparticles. AMPs open up many new beneficial possibilities in treating severe and deadly infections and even malignant diseases. It is precisely this knowledge that is increasingly growing about yet undiscovered immune functions. It is also essential in discovering other possibilities of the human genome in creating a more comfortable, good quality, and longer life.
References
- Hendry, A.P.; Kinnison, M.T.; Heino, M.; Day, T.; Smith, T.B.; Fitt, G.; Bergstrom, C.T.; Oakeshott, J.; Jørgensen, P.S.; Zalucki, M.P.; et al. Evolutionary principles and their practical application. Evol. Appl. 2011, 4, 159–183.
- Van Baarlen, P.; Van Belkum, A.; Summerbell, R.C.; Crous, P.W.; Thomma, B.P.H.J. Molecular mechanisms of pathogenicity: How do pathogenic microorganisms develop cross-kingdom host jumps? FEMS Microbiol. Rev. 2007, 31, 239–277.
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318.
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658.
- Tenover, F.C. Mechanisms of Antimicrobial Resistance in Bacteria. Am. J. Med. 2006, 34, S3–S10.
- Patini, R.; Mangino, G.; Martellacci, L.; Quaranta, G.; Masucci, L.; Gallenzi, P. The effect of different antibiotic regimens on bacterial resistance: A systematic review. Antibiotics 2020, 9, 22. [Google Scholar] [CrossRef]
- Landecker, H. Antibiotic Resistance and the Biology of History. Body Soc. 2016, 22, 19–52.
- Hay, S.I.; Rao, P.C.; Dolecek, C.; Day, N.P.J.; Stergachis, A.; Lopez, A.D.; Murray, C.J.L. Measuring and mapping the global burden of antimicrobial resistance. BMC Med. 2018, 16, 78.
- Gajdács, M.; Albericio, F. Antibiotic resistance: From the bench to patients. Antibiotics 2019, 8, 129.
- Makowski, M.; Silva, Í.C.; Do Amaral, C.P.; Gonçalves, S.; Santos, N.C. Advances in lipid and metal nanoparticles for antimicrobial peptide delivery. Pharmaceutics 2019, 11, 588.
- Teixeira, M.C.; Carbone, C.; Sousa, M.C.; Espina, M.; Garcia, M.L.; Sanchez-Lopez, E.; Souto, E.B. Nanomedicines for the delivery of antimicrobial peptides (Amps). Nanomaterials 2020, 10, 560.
- Mohanty, S.; Jena, P.; Mehta, R.; Pati, R.; Banerjee, B.; Patil, S.; Sonawane, A. Cationic antimicrobial peptides and biogenic silver nanoparticles kill mycobacteria without eliciting dna damage and cytotoxicity in mouse macrophages. Antimicrob. Agents Chemother. 2013, 57, 3688–3698.
- Gao, J.; Na, H.; Zhong, R.; Yuan, M.; Guo, J.; Zhao, L.; Wang, Y.; Wang, L.; Zhang, F. One step synthesis of antimicrobial peptide protected silver nanoparticles: The core-shell mutual enhancement of antibacterial activity. Colloids Surf. B Biointerfaces 2020, 186, 110704.
- Mukhopadhyay, S.; Bharath Prasad, A.S.; Mehta, C.H.; Nayak, U.Y. Antimicrobial peptide polymers: No escape to ESKAPE pathogens—A review. World J. Microbiol. Biotechnol. 2020, 36, 131–132.
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539.
- Jamal, M.; Tasneem, U.; Hussain, T.; Andleeb, S. Bacterial Biofilm: Its Composition, Formation and Role in Human Infections. Res. Rev. J. Microbiol. Biotechnol. 2015, 4. Available online: https://www.rroij.com/open-access/bacterial-biofilm-its-composition-formation-and-role-in-human-infections.pdf (accessed on 10 October 2020).
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824.
- Bayramov, D.F.; Neff, J.A. Beyond conventional antibiotics—New directions for combination products to combat biofilm. Adv. Drug Deliv. Rev. 2017, 112, 48–60.
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-Molecule Inhibition of Bacterial Biofilm. ASC Omega 2020, 5, 3108–3115.
- Patini, R.; Cattani, P.; Marchetti, S.; Isola, G.; Quaranta, G.; Gallenzi, P. Evaluation of predation capability of periodontopathogens bacteria by Bdellovibrio Bacteriovorus HD100. An in Vitro Study. Materials 2019, 12, 9.
- Annous, B.A.; Fratamico, P.M.; Smith, J.L. Scientific status summary: Quorum sensing in biofilms: Why bacteria behave the way they do. J. Food Sci. 2009, 74, R24–R37.
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554.
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial quorum sensing and microbial community interactions. MBio 2018, 9.
- Wille, J.; Coenye, T. Biofilm dispersion: The key to biofilm eradication or opening Pandora’s box? Biofilm 2020, 2, 100027.
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886.
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352.
- Bondy-Denomy, J.; Davidson, A.R. When a virus is not a parasite: The beneficial effects of prophages on bacterial fitness. J. Microbiol. 2014, 52, 235–242.
- Fernández, L.; Rodríguez, A.; García, P. Phage or foe: An insight into the impact of viral predation on microbial communities. ISME J. 2018, 12, 1171–1179.
- Talapko, J.; Škrlec, I.; Alebić, T.; Bekić, S.; Včev, A. From Bacteriophage to Antibiotics and Back. Coll. Antropol. 2018, 42, 131–138.
- Dakheel, K.H.; Rahim, R.A.; Neela, V.K.; Al-Obaidi, J.R.; Hun, T.G.; Isa, M.N.M.; Yusoff, K. Genomic analyses of two novel biofilm-degrading methicillin-resistant Staphylococcus aureus phages. BMC Microbiol. 2019, 19, 114.
- Lewenza, S. Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front. Microbiol. 2013, 4, 21.
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4.
- Hansen, M.F.; Svenningsen, S.L.; Røder, H.L.; Middelboe, M.; Burmølle, M. Big Impact of the Tiny: Bacteriophage—Bacteria Interactions in Biofilms. Trends Microbiol. 2019, 27, 739–752.
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G.M. Formation of biofilms under phage predation: Considerations concerning a biofilm increase. Biofouling 2013, 29, 457–468.
- Milho, C.; Andrade, M.; Vilas Boas, D.; Alves, D.; Sillankorva, S. Antimicrobial assessment of phage therapy using a porcine model of biofilm infection. Int. J. Pharm. 2019, 557, 112–123.
- Tait, K.; Skillman, L.C.; Sutherland, I.W. The efficacy of bacteriophage as a method of biofilm eradication. Biofouling 2002, 18, 305–311.
- Nobrega, F.L.; Costa, A.R.; Kluskens, L.D.; Azeredo, J. Revisiting phage therapy: New applications for old resources. Trends Microbiol. 2015, 23, 185–191.
- Ferriol-González, C.; Domingo-Calap, P. Phages for biofilm removal. Antibiotics 2020, 9, 268.
- Bull, J.; Christensen, K.; Scott, C.; Jack, B.; Crandall, C.; Krone, S. Phage-Bacterial Dynamics with Spatial Structure: Self Organization around Phage Sinks Can Promote Increased Cell Densities. Antibiotics 2018, 7, 8.
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931.
- Penadés, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178.
- Torres-Barceló, C.; Hochberg, M.E. Evolutionary Rationale for Phages as Complements of Antibiotics. Trends Microbiol. 2016, 24, 249–256.
- Moelling, K.; Broecker, F.; Willy, C. A Wake-Up Call: We Need Phage Therapy Now. Viruses 2018, 10, 688.
- Baker, S.J.; Payne, D.J.; Rappuoli, R.; De Gregorio, E. Technologies to address antimicrobial resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 12887–12895.
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511.
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194.
- Bahar, A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575.
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4.
- Borah, A.; Deb, B.; Chakraborty, S. A Crosstalk on Antimicrobial Peptides. Int. J. Pept. Res. Ther. 2020, 1–16.
- Zharkova, M.S.; Orlov, D.S.; Golubeva, O.Y.; Chakchir, O.B.; Eliseev, I.E.; Grinchuk, T.M.; Shamova, O.V. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics-a novel way to combat antibiotic resistance? Front. Cell. Infect. Microbiol. 2019, 9, 128.
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55.
- Yang, D.; Biragyn, A.; Hoover, D.M.; Lubkowski, J.; Oppenheim, J.J. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 2004, 22, 181–215.
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395.
- Carmona-Ribeiro, A.; de Melo Carrasco, L. Novel Formulations for Antimicrobial Peptides. Int. J. Mol. Sci. 2014, 15, 18040–18083.
- Vilas Boas, L.C.P.; Campos, M.L.; Berlanda, R.L.A.; de Carvalho Neves, N.; Franco, O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542.
- Bechinger, B.; Gorr, S.U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260.
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 2016, 26, 43–57.
- Lee, T.-H.; Hall, K.N.; Aguilar, M.-I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2015, 16, 25–39.
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547.
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334.
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720.
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704.
- Kang, J.; Dietz, M.J.; Li, B. Antimicrobial peptide LL-37 is bactericidal against Staphylococcus aureus biofilms. PLoS ONE 2019, 14, e0216676.
- Hilchie, A.L.; Wuerth, K.; Hancock, R.E.W. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 2013, 9, 761–768.
- Fox, J.L. Antimicrobial peptides stage a comeback. Nat. Biotechnol. 2013, 31, 379–382.
- Vaara, M. New approaches in peptide antibiotics. Curr. Opin. Pharmacol. 2009, 9, 571–576.
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141.
- Pinheiro Da Silva, F.; MacHado, M.C.C. Antimicrobial peptides: Clinical relevance and therapeutic implications. Peptides 2012, 36, 308–314.
- Da Cunha, N.B.; Cobacho, N.B.; Viana, J.F.C.; Lima, L.A.; Sampaio, K.B.O.; Dohms, S.S.M.; Ferreira, A.C.R.; de la Fuente-Núñez, C.; Costa, F.F.; Franco, O.L.; et al. The next generation of antimicrobial peptides (AMPs) as molecular therapeutic tools for the treatment of diseases with social and economic impacts. Drug Discov. Today 2017, 22, 234–248.
- Divyashree, M.; Mani, M.K.; Reddy, D.; Kumavath, R.; Ghosh, P.; Azevedo, V.; Barh, D. Clinical Applications of Antimicrobial Peptides (AMPs): Where Do We Stand Now? Protein Pept. Lett. 2019, 27, 120–134.
- Lei, J.; Sun, L.C.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q.Y. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931.
- Lemon, D.J.; Kay, M.K.; Titus, J.K.; Ford, A.A.; Chen, W.; Hamlin, N.J.; Hwang, Y.Y. Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J. Microbiol. 2019, 57, 532–538.
- Reinhardt, A.; Neundorf, I. Design and application of antimicrobial peptide conjugates. Int. J. Mol. Sci. 2016, 17, 701.
- Namivandi-Zangeneh, R.; Sadrearhami, Z.; Dutta, D.; Willcox, M.; Wong, E.H.H.; Boyer, C. Synergy between Synthetic Antimicrobial Polymer and Antibiotics: A Promising Platform to Combat Multidrug-Resistant Bacteria. ACS Infect. Dis. 2019, 5, 1357–1365.
- Holban, A.M.; Gestal, M.C.; Grumezescu, A.M. Control of biofilm-associated infections by signaling molecules and nanoparticles. Int. J. Pharm. 2016, 510, 409–418.
- Biswaro, L.S.; Sousa, M.G.d.C.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 2018, 9, 855.
- Pelling, H.; Nzakizwanayo, J.; Milo, S.; Denham, E.L.; MacFarlane, W.M.; Bock, L.J.; Sutton, J.M.; Jones, B.V. Bacterial biofilm formation on indwelling urethral catheters. Lett. Appl. Microbiol. 2019, 68, 277–293.
- Talapko, J.; Matijević, T.; Juzbašić, M.; Antolović-Požgain, A.; Škrlec, I. Antibacterial Activity of Silver and Its Application in Dentistry, Cardiology and Dermatology. Microorganisms 2020, 8, 1400.
- Ramesh, S.; Grijalva, M.; Debut, A.; De La Torre, B.G.; Albericio, F.; Cumbal, L.H. Peptides conjugated to silver nanoparticles in biomedicine-a “value-added” phenomenon. Biomater. Sci. 2016, 4, 1713–1725.
- Almaaytah, A.; Mohammed, G.; Abualhaijaa, A.; Al-Balas, Q. Development of novel ultrashort antimicrobial peptide nanoparticles with potent antimicrobial and antibiofilm activities against multidrug-resistant bacteria. Drug Des. Devel. Ther. 2017, 11, 3159–3170.
- Pal, I.; Bhattacharyya, D.; Kar, R.K.; Zarena, D.; Bhunia, A.; Atreya, H.S. A Peptide-Nanoparticle System with Improved Efficacy against Multidrug Resistant Bacteria. Sci. Rep. 2019, 9, 1–11.
- Chaudhari, A.A.; Ashmore, D.; Nath, S.D.; Kate, K.; Dennis, V.; Singh, S.R.; Owen, D.R.; Palazzo, C.; Arnold, R.D.; Miller, M.E.; et al. A novel covalent approach to bio-conjugate silver coated single walled carbon nanotubes with antimicrobial peptide. J. Nanobiotechnol. 2016, 14.
- Gakiya-Teruya, M.; Palomino-Marcelo, L.; Pierce, S.; Angeles-Boza, A.M.; Krishna, V.; Rodriguez-Reyes, J.C.F. Enhanced antimicrobial activity of silver nanoparticles conjugated with synthetic peptide by click chemistry. J. Nanopart. Res. 2020, 22, 90.
- Ruden, S.; Hilpert, K.; Berditsch, M.; Wadhwani, P.; Ulrich, A.S. Synergistic interaction between silver nanoparticles and membrane-permeabilizing antimicrobial peptides. Antimicrob. Agents Chemother. 2009, 53, 3538–3540.
