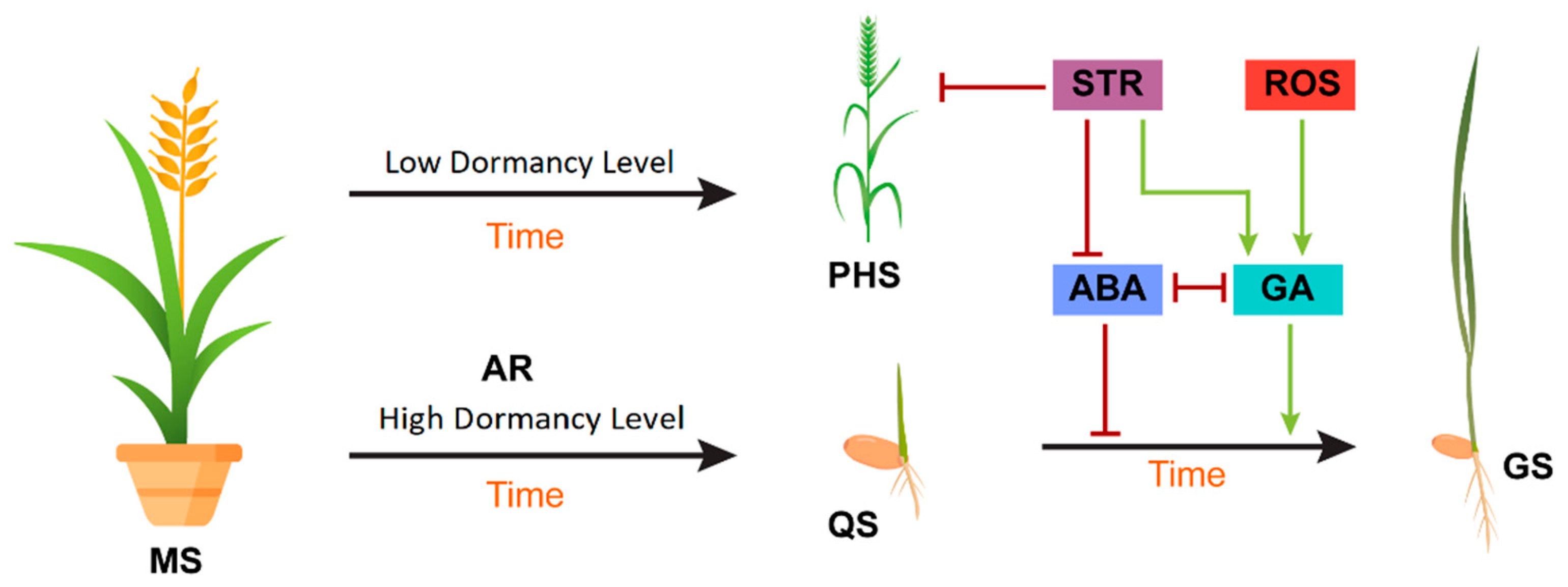
With the burgeoning population of the world, the successful germination of seeds to achieve maximum crop production is very important. Seed germination is a precise balance of phytohormones, light, and temperature that induces endosperm decay. Abscisic acid and gibberellins—mainly with auxins, ethylene, and jasmonic and salicylic acid through interdependent molecular pathways—lead to the rupture of the seed testa, after which the radicle protrudes out and the endosperm provides nutrients according to its growing energy demand. The incident light wavelength and low and supra-optimal temperature modulates phytohormone signaling pathways that induce the synthesis of ROS, which results in the maintenance of seed dormancy and germination.
- seed germination and dormancy
- phytohormones
- light
- temperature and endosperm decay
1. Introduction
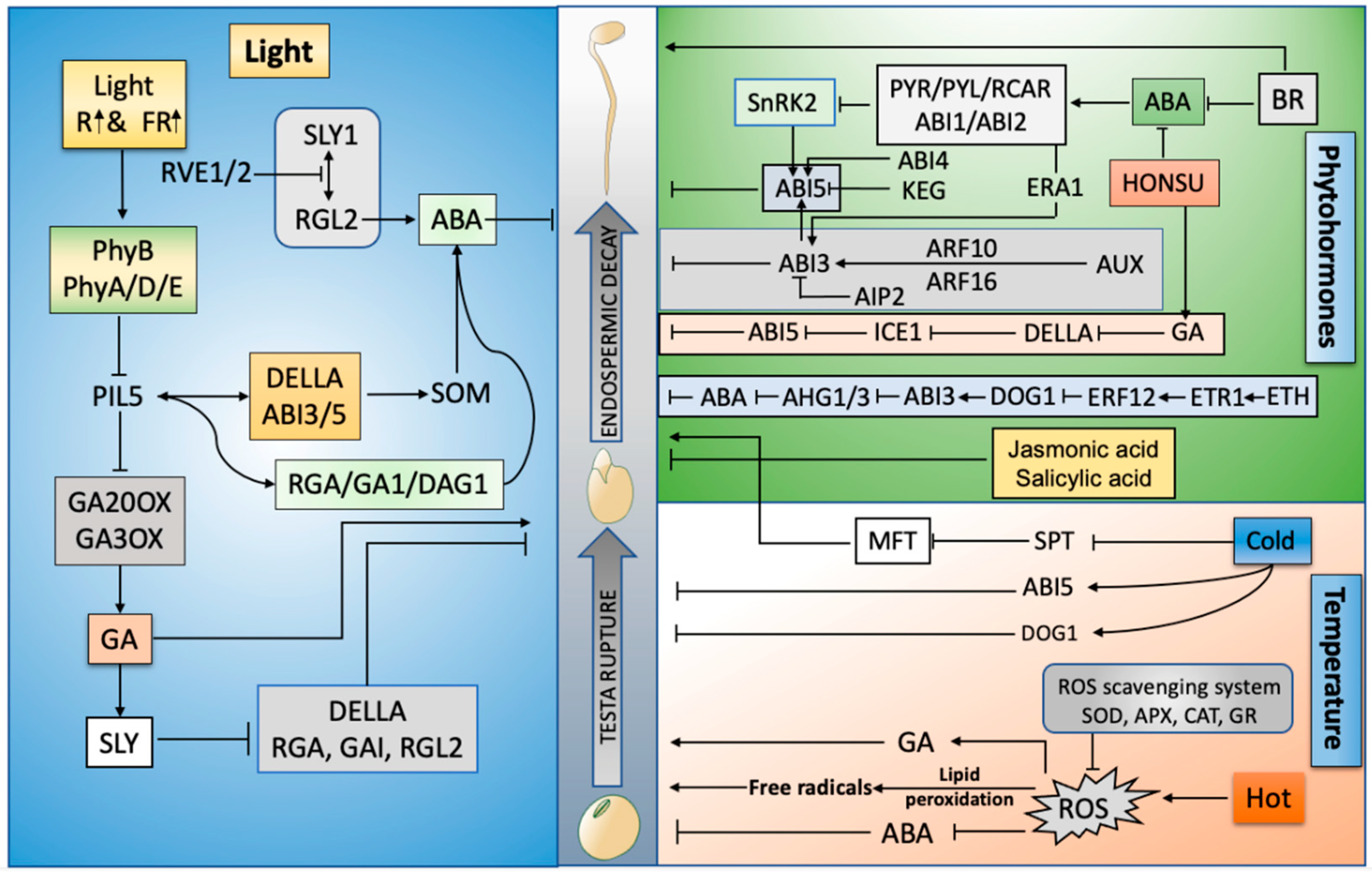
2. Phytohormone Regulation of Dormancy and Germination
| Name of Gene | Mutant Dormancy Level | General Description of Gene | References |
|---|---|---|---|
| ABI3 | Decreased | Positively regulates ABA signaling and represses seed germination | [27] |
| ABI4 | Decreased | Positively regulates ABA signaling and represses seed germination | [28,29][28][29] |
| ABI5 | Not Changed | Positively regulates ABA signaling and represses seed germination | [30,31][30][31] |
| NCED5 | Decreased | ABA-biosynthesis gene; the ABA content is decreased | [32] |
| CYP707A1/2 | Enhanced | ABI4 negatively regulates its transcription | [33] |
| GAI/2 | Enhanced | GA-biosynthesis genes; GA content is decreased in mutants | [23] |
| GA2oxs | Decreased | GA-inactivate genes; GA content is upregulated in mutants | [34] |
| RGL2/SPY | Enhanced | GA signaling is blocked in mutants | [35] |
| MYB96 | Decreased | Decreases transcription of ABI4 and some ABA biogenesis genes | [36,[3637]][37] |
| DOG1 | Enhanced | ABA sensitivity of dog1 seeds is unchanged | [38,39][38][39] |
| SUVH4/SUVH5 | Enhanced | Repress DOG1 and ABI3 transcription | [40] |
| LDL1/LDL2 | Enhanced | Repress seed dormancy by negatively regulating DOG1 | [17] |
| WRKY41 | Decreased | WRKY41 directly promotes ABI3 transcription | [41] |
| RAF10/RAF11 | Decreased | Directly enhances ABI3 transcription | [42] |
| DEP | Decreased | Promotes ABI3 transcription | [43] |
| SPT | Decreased in Ler but enhanced in Col background | Opposite roles in Ler and Col ecotypes | [44,[4445]][45] |
| ARF10/ARF16 | Decreased | ARF10/ARF16 directly promote ABI3 transcription | [46] |
| BIN2 | Not mentioned | Phosphorylates and stabilizes ABI5 to enhance ABA signaling | [47] |
| PKS5 | Not mentioned | Phosphorylates ABI5 (Ser42) and controls transcription of ABA-responsive genes | [48] |
| HONSU | Enhanced | A PP2C protein that impairs ABA signaling | [20] |
| RDO5 | Enhanced | Its ABA sensitivity and content remain unchanged | [21] |
| ABI1/2 | Decreased | Dominant-negative mutants; the mutated proteins cannot interact with ABA receptors | [49] |
| CHO1 | Decreased | Acts upstream on ABI4 genetically | [50] |
| OsAP2-39 | Decreased | Promotes OsNCEDI and OsEUI, thereby enhancing ABA biogenesis and impairing GA accumulation | [51] |
| DDF1 | Decreased | Directly promotes GAox7 and thus decreases GA content | [52] |
3. Light Controls Seed Germination and Dormancy
References
- Carrera-Castaño, G.; Calleja-Cabrera, J.; Pernas, M.; Gómez, L.; Oñate-Sánchez, L. An Updated Overview on the Regulation of Seed Germination. Plants 2020, 9, 703.
- Wang, H.; Zhao, K.; Li, X.; Chen, X.; Liu, W.; Wang, J. Factors affecting seed germination and emergence of Aegilops tauschii. Weed Res. 2020, 60, 171–181.
- Oracz, K.; Karpiński, S. Phytohormones signaling pathways and ROS involvement in seed germination. Front. Plant Sci. 2016, 7, 864.
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.-H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45.
- Wang, Y.; Htwe, Y.M.; Li, J.; Shi, P.; Zhang, D.; Zhao, Z.; Ihase, L.O. Integrative omics analysis on phytohormones involved in oil palm seed germination. BMC Plant Biol. 2019, 19, 1–14.
- MacGregor, D.R.; Kendall, S.L.; Florance, H.; Fedi, F.; Moore, K.; Paszkiewicz, K.; Smirnoff, N.; Penfield, S. Seed production temperature regulation of primary dormancy occurs through control of seed coat phenylpropanoid metabolism. N. Phytol. 2015, 205, 642–652.
- Farooq, M.A.; Zhang, X.; Zafar, M.M.; Ma, W.; Zhao, J. Roles of Reactive Oxygen Species and Mitochondria in Seed Germination. Front. Plant Sci. 2021, 12, 781734.
- Xia, Q.; Ponnaiah, M.; Cueff, G.; Rajjou, L.; Prodhomme, D.; Gibon, Y.; Bailly, C.; Corbineau, F.; Meimoun, P.; El-Maarouf-Bouteau, H. Integrating proteomics and enzymatic profiling to decipher seed metabolism affected by temperature in seed dormancy and germination. Plant Sci. 2018, 269, 118–125.
- Oh, E.; Kim, J.; Park, E.; Kim, J.-I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058.
- He, H.; Willems, L.A.J.; Batushansky, A.; Fait, A.; Hanson, J.; Nijveen, H.; Hilhorst, H.; Bentsink, L. Effects of Parental Temperature and Nitrate on Seed Performance are Reflected by Partly Overlapping Genetic and Metabolic Pathways. Plant Cell Physiol. 2016, 57, 473–487.
- Meng, Y.; Shuai, H.; Luo, X.; Chen, F.; Zhou, W.; Yang, W.; Shu, K. Karrikins: Regulators involved in phytohormone signaling networks during seed germination and seedling development. Front. Plant Sci. 2017, 7, 2021.
- Springthorpe, V.; Penfield, S. Flowering time and seed dormancy control use external coincidence to generate life history strategy. Elife 2015, 4, e05557.
- Jiang, A.; Guo, Z.; Pan, J.; Yang, Y.; Zhuang, Y.; Zuo, D.; Hao, C.; Gao, Z.; Xin, P.; Chu, J.; et al. The PIF1-miR408-PLANTACYANIN repression cascade regulates light-dependent seed germination. Plant Cell 2021, 33, 1506–1529.
- Yang, L.-W.; Liu, S.; Lin, R. The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol. 2020, 62, 1310–1326.
- Zhang, K.; Yao, L.; Zhang, Y.; Baskin, J.M.; Baskin, C.C.; Xiong, Z.; Tao, J. A review of the seed biology of Paeonia species (Paeoniaceae), with particular reference to dormancy and germination. Planta 2019, 249, 291–303.
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121.
- Zhou, S.F.; Sun, L.; Valdés, A.E.; Engström, P.; Song, Z.T.; Lu, S.J.; Liu, J.X. Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in Arabidopsis. New Phytol. 2015, 208, 188–197.
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708.
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679.
- Kim, W.; Lee, Y.; Park, J.; Lee, N.; Choi, G. HONSU, a Protein Phosphatase 2C, Regulates Seed Dormancy by Inhibiting ABA Signaling in Arabidopsis. Plant Cell Physiol. 2013, 54, 555–572.
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375.
- Zhang, W.; Qu, L.-W.; Zhao, J.; Xue, L.; Dai, H.-P.; Xing, G.-M.; Lei, J.-J. Practical Methods for Breaking Seed Dormancy in a Wild Ornamental Tulip Species Tulipa thianschanica Regel. Agronomy 2020, 10, 1765.
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002, 16, 646–658.
- Resentini, F.; Felipo-Benavent, A.; Colombo, L.; Blázquez, M.A.; Alabadí, D.; Masiero, S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 2015, 8, 482–485.
- Steber, C.M.; Cooney, S.E.; McCourt, P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 1998, 149, 509–521.
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C. ABI 4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361.
- Finkelstein, R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994, 5, 765–771.
- Brocard-Gifford, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulatory Networks in Seeds Integrating Developmental, Abscisic Acid, Sugar, and Light Signaling. Plant Physiol. 2003, 131, 78–92.
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular Aspects of Seed Dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415.
- Kong, D.; Ju, C.; Parihar, A.; Kim, S.; Cho, D.; Kwak, J.M. Arabidopsis Glutamate Receptor Homolog3.5 Modulates Cytosolic Ca2+ Level to Counteract Effect of Abscisic Acid in Seed Germination. Plant Physiol. 2015, 167, 1630–1642.
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 Regulates Primary Seed Dormancy by Regulating the Biogenesis of Abscisic Acid and Gibberellins in Arabidopsis. PLoS Genet. 2013, 9, e1003577.
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512.
- Matakiadis, T.; Alboresi, A.; Jikumaru, Y.; Tatematsu, K.; Pichon, O.; Renou, J.-P.; Kamiya, Y.; Nambara, E.; Truong, H.-N. The Arabidopsis Abscisic Acid Catabolic Gene CYP707A2 Plays a Key Role in Nitrate Control of Seed Dormancy. Plant Physiol. 2008, 149, 949–960.
- Yamauchi, Y.; Takeda-Kamiya, N.; Hanada, A.; Ogawa, M.; Kuwahara, A.; Seo, M.; Kamiya, Y.; Yamaguchi, S. Contribution of Gibberellin Deactivation by AtGA2ox2 to the Suppression of Germination of Dark-Imbibed Arabidopsis thaliana Seeds. Plant Cell Physiol. 2007, 48, 555–561.
- Jacobsen, S.E.; Olszewski, N.E. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 1993, 5, 887–896.
- Lee, H.G.; Lee, K.; Seo, P.J. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 2015, 87, 371–381.
- Lee, K.; Lee, H.G.; Yoon, S.; Kim, H.U.; Seo, P.J. The Arabidopsis MYB96 Transcription Factor Is a Positive Regulator of ABSCISIC ACID-INSENSITIVE4 in the Control of Seed Germination. Plant Physiol. 2015, 168, 677–689.
- Nakabayashi, K.; Bartsch, M.; Xiang, Y.; Miatton, E.; Pellengahr, S.; Yano, R.; Seo, M.; Soppe, W.J. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 2012, 24, 2826–2838.
- Graeber, K.; Linkies, A.; Steinbrecher, T.; Mummenhoff, K.; Tarkowská, D.; Turečková, V.; Ignatz, M.; Sperber, K.; Voegele, A.; De Jong, H. DELAY OF GERMI-NATION 1 mediates a conserved coat-dormancy mechanism for the temperature-and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 2014, 111, E3571–E3580.
- Zheng, J.; Chen, F.; Wang, Z.; Cao, H.; Li, X.; Deng, X.; Soppe, W.J.; Li, Y.; Liu, Y. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616.
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823.
- Lee, S.-J.; Lee, M.H.; Kim, J.-I.; Kim, S.Y. Arabidopsis Putative MAP Kinase Kinase Kinases Raf10 and Raf11 are Positive Regulators of Seed Dormancy and ABA Response. Plant Cell Physiol. 2014, 56, 84–97.
- Barrero, J.M.; Millar, A.A.; Griffiths, J.; Czechowski, T.; Scheible, W.R.; Udvardi, M.; Reid, J.B.; Ross, J.J.; Jacobsen, J.V.; Gubler, F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010, 61, 611–622.
- Vaistij, F.E.; Gan, Y.; Penfield, S.; Gilday, A.D.; Dave, A.; He, Z.; Josse, E.-M.; Choi, G.; Halliday, K.J.; Graham, I.A. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc. Natl. Acad. Sci. USA 2013, 110, 10866–10871.
- Belmonte, M.F.; Kirkbride, R.C.; Stone, S.L.; Pelletier, J.M.; Bui, A.Q.; Yeung, E.C.; Hashimoto, M.; Fei, J.; Harada, C.M.; Munoz, M.D.; et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 2013, 110, E435–E444.
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z.-H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490.
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE2 interacts with AB-SCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408.
- Zhou, X.; Hao, H.; Zhang, Y.; Bai, Y.; Zhu, W.; Qin, Y.; Yuan, F.; Zhao, F.; Wang, M.; Hu, J. PKS5/CIPK11, a SnRK3-type protein kinase, is important for ABA responses in Arabidopsis through phosphorylation of ABI5. Plant Physiol. 2015.
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071.
- Yano, R.; Kanno, Y.; Jikumaru, Y.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 2009, 151, 641–654.
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.-M.; Rothstein, S.J. The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice. PLoS Genet. 2010, 6, e1001098.
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626.
- Kerchev, P.I.; Pellny, T.K.; Vivancos, P.D.; Kiddle, G.; Hedden, P.; Driscoll, S.; Vanacker, H.; Verrier, P.; Hancock, R.D.; Foyer, C.H. The Transcription Factor ABI4 Is Required for the Ascorbic Acid–Dependent Regulation of Growth and Regulation of Jasmonate-Dependent Defense Signaling Pathways in Arabidopsis. Plant Cell 2011, 23, 3319–3334.
- Shuai, H.; Meng, Y.; Luo, X.; Chen, F.; Zhou, W.; Dai, Y.; Qi, Y.; Du, J.; Yang, F.; Liu, J. Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci. Rep. 2017, 7, 12620.
- Majid, A.; Mohsen, S.; Mandana, A.; Saeid, J.-H.; Ezatollah, E.; Fariborz, S. Biochemistry, The effects of different levels of salinity and indole-3-acetic acid (IAA) on early growth and germination of wheat seedling. J. Stress Physiol. Biochem. 2013, 9, 329–338.
- Liu, A.; Gao, F.; Kanno, Y.; Jordan, M.C.; Kamiya, Y.; Seo, M.; Ayele, B.T. Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE 2013, 8, e56570.
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539.
- Jiroutova, P.; Oklestkova, J.; Strnad, M. Crosstalk between brassinosteroids and ethylene during plant growth and under abiotic stress conditions. Int. J. Mol. Sci. 2018, 19, 3283.
- Yanik, F.; Aytürk, Ö.; Genc, A.C.; Vardar, F. Salicylic acid-induced germination, biochemical and developmental alterations in rye (Secale cereale L.). Acta Bot. Croat. 2018, 77, 45–50.
- Tuan, P.A.; Yamasaki, Y.; Kanno, Y.; Seo, M.; Ayele, B.T. Transcriptomics of cytokinin and auxin metabolism and signaling genes during seed maturation in dormant and non-dormant wheat genotypes. Sci. Rep. 2019, 9, 1–16.
- Linkies, A.; Leubner-Metzger, G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2012, 31, 253–270.
- Fantini, E.; Facella, P. Cryptochromes in the field: How blue light influences crop development. Physiol. Plant. 2020, 169, 336–346.
- Barrero, J.M.; Downie, A.B.; Xu, Q.; Gubler, F. A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 2014, 26, 1094–1104.
- Hajiabbasi, M.; Afshari, R.T.; Abbasi, A.; Seyyedi, S. Germination and gene expression as affected by aminocyclopropane-1-carboxylic acid in deteriorated soybean seed. J. Crop Improv. 2020, 34, 505–517.
- Lee, K.P.; Piskurewicz, U.; Turečková, V.; Carat, S.; Chappuis, R.; Strnad, M.; Fankhauser, C.; Lopez-Molina, L. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012, 26, 1984–1996.
- Hennig, L.; Stoddart, W.M.; Dieterle, M.; Whitelam, G.C.; Schäfer, E. Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol. 2002, 128, 194–200.
- Vaistij, F.E.; Barros-Galvão, T.; Cole, A.F.; Gilday, A.D.; He, Z.; Li, Y.; Harvey, D.; Larson, T.R.; Graham, I.A. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, 8442–8447.
- Park, E.; Kim, Y.; Choi, G. Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 2018, 30, 1277–1292.
- Béziat, C.; Barbez, E.; Feraru, M.I.; Lucyshyn, D.; Kleine-Vehn, J. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat. Plants 2017, 3, 17105.
- Yavari, N. An Integrated Approach to Unravel the Physiological and Molecular Basis of Arabidopsis Thaliana Response to Narrow-Wavelength Light; McGill University: Montréal, QC, Canada, 2020.
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476.
- De Wit, M.; Keuskamp, D.H.; Bongers, F.J.; Hornitschek, P.; Gommers, C.M.; Reinen, E.; Martínez-Cerón, C.; Fankhauser, C.; Pierik, R. Integration of Phytochrome and Cryptochrome Signals Determines Plant Growth during Competition for Light. Curr. Biol. 2016, 26, 3320–3326.
- Lymperopoulos, P.; Msanne, J.; Rabara, R. Phytochrome and phytohormones: Working in tandem for plant growth and development. Front. Plant Sci. 2018, 9, 1037.
