The importance of mitochondria in mammalian cells is widely known. Several biochemical reactions and pathways take place within mitochondria: among them, there are those involving the biogenesis of the iron–sulfur (Fe-S) clusters. The latter are evolutionarily conserved, ubiquitous inorganic cofactors, performing a variety of functions, such as electron transport, enzymatic catalysis, DNA maintenance, and gene expression regulation. The synthesis and distribution of Fe-S clusters are strictly controlled cellular processes that involve several mitochondrial proteins that specifically interact each other to form a complex machinery (Iron Sulfur Cluster assembly machinery, ISC machinery hereafter). This machinery ensures the correct assembly of both [2Fe-2S] and [4Fe-4S] clusters and their insertion in the mitochondrial target proteins.
- iron–sulfur cluster
- mitochondrial proteins
- multiple mitochondrial dysfunction syndromes
- rare diseases
1. Introduction
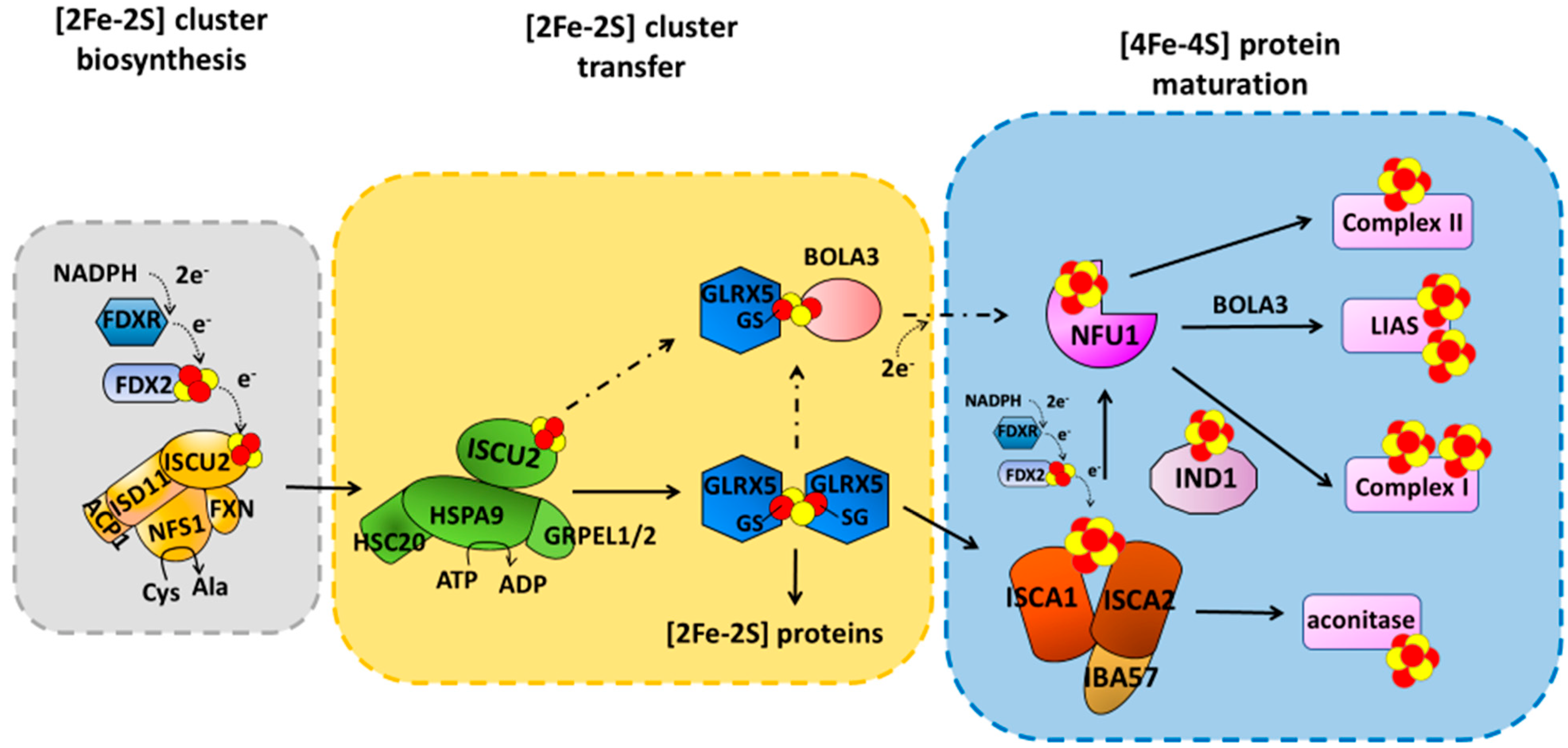
2. [4Fe-4S] Cluster Assembly in Mitochondria
The mechanism responsible for the maturation of mitochondrial [4Fe-4S] proteins is not yet fully defined. Indeed, both transient protein–protein interactions and stable protein complexes involved in such processes are still only partially characterized at the molecular level, as well as the picture of the operative protein–protein interaction network is still argument of debate in the literature, where two main models have been proposed. One model proposed that NFU1 operates with ISCU2 and ISCA1 proteins to assemble a [4Fe-4S]2+ cluster [25]. Specifically, the latter proteins provide a [2Fe-2S]2+ cluster each to NFU1 forming a ISCU2-ISCA1-NFU1 complex; this ternary complex receives two electrons from FDX2 to assemble a [4Fe-4S]2+ cluster on dimeric NFU1 [25]. The [4Fe-4S]2+ cluster assembled on NFU1 was proposed to be then transferred to apo recipient proteins [25]. However, at the moment, the most accredited model, based on experimental data collected by different research groups, supports a different protein–protein interaction network, described hereafter. This model has been here considered in the description of the effects that pathogenic mutations have on the maturation of mitochondrial [4Fe-4S] proteins. ISCA1 and ISCA2 preferentially form a stable hetero-dimeric complex responsible for assembling a [4Fe-4S] cluster [21,26][21][26]. The latter is assembled through a reductive coupling of two [2Fe-2S] clusters that are donated to the apo ISCA1-ISCA2 hetero-complex by homo-dimeric [2Fe-2S]2+ GLRX5 (Figure 1) [21,22,27][21][22][27]. The latter bridges a [2Fe-2S]2+ cluster coordinated by two glutathione molecules and a conserved cysteine per each subunit of the homodimer, and transiently interacts with ISCA1 and ISCA2 to transfer the two [2Fe-2S]2+ clusters cargo to the apo ISCA1-ISCA2 hetero-complex via sequential molecular events [21,22][21][22]. Electrons required to couple the two received [2Fe-2S]2+ clusters are donated by FDX2 (Figure 1) [27], but no information is yet available on which is the specific electron acceptor. IBA57 has been shown to be required to assemble the [4Fe-4S]2+ cluster on ISCA1-ISCA2 complex, but its specific molecular function is still not clarified. IBA57 was shown to form a hetero-dimeric complex with ISCA2, but not with ISCA1, via bridging a [2Fe-2S] cluster [28,29][28][29]. Considering that the ISCA2-IBA57 complex can stabilize both reduced [2Fe-2S]+ and oxidized [2Fe-2S]2+ bound clusters [28], a possibility is that the [2Fe-2S] IBA57-ISCA2 heterodimeric complex is the entry point of the electrons donated by FDX2, but this still needs experimental evidence. The assembled [4Fe-4S]2+ cluster can then be inserted into apo recipient proteins, such as aconitase, without the requirement of further accessory proteins [6,30,31][6][30][31] or can be transferred to NFU1 or IND1 that then mediate the [4Fe-4S]2+ cluster insertion specifically into recipient proteins such as complex I, complex II and lipoyl synthase (LIAS) (Figure 1) [6,30,31][6][30][31]. While the molecular mechanism involving IND1 in the insertion of the [4Fe-4S]2+ cluster into complex I is not yet characterized, the insertion of the [4Fe-4S]2+ cluster into LIAS has been deeply investigated. Sequential protein–protein interactions involving ISCA1 and NFU1 have been shown to be responsible for the insertion of the [4Fe-4S] cluster into LIAS [32,33][32][33]. LIAS is a member of the radical S-adenosylmethionine (SAM) superfamily [34,35][34][35]. It catalyzes the final step of the biosynthesis of lipoyl cofactor [36,37,38][36][37][38] and binds two [4Fe-4S] clusters [39,40][39][40]: a [4Fe-4S] cluster (FeSRS), typical of all radical SAM enzymes, and a [4Fe-4S] cluster (FeSaux) that provides two sulfur atoms to the lipoyl cofactor [41,42][41][42]. It has been shown that the C-domain of NFU1 drives first [4Fe-4S]2+ cluster delivery from the ISCA1-ISCA2 complex, where the [4Fe-4S]2+ cluster is assembled, to a [4Fe-4S]2+ ISCA1-NFU1 intermediate complex, which then specifically directs the cluster into the FeSRS site of LIAS [32,33][32][33]. According to this molecular function, NFU1 has been shown to form two hetero-complexes with ISCA1 and LIAS [32,33][32][33]. These two complexes have been recently characterized at a molecular level, and it has been shown that, in both cases, only the C-domain of NFU1 is involved in the protein–protein interaction [32,33][32][33]. This domain therefore drives the assembled [4Fe-4S]2+ cluster from the ISCA1-ISCA2 complex to the final destination. BOLA3 has been implicated in the latter transfer step to LIAS (Figure 1) on the basis of clinical phenotypes of patients with pathogenic variants in the BOLA3 gene similar to those of patients with pathogenic variants in the NFU1 gene [43]. However, how BOLA3 contributes to this process is yet not defined. Indeed, in vitro studies showed that NFU1 and BOLA3 do not interact with each other in various experimental conditions, i.e., BOLA3 with either apo or [4Fe-4S]2+ NFU1 [20,44][20][44]. The only well-defined partner of BOLA3 is GLRX5, which operates upstream in the [4Fe-4S] cluster maturation pathway as a [2Fe-2S] cluster donor. GLRX5 forms an apo hetero-dimeric complex with BOLA3 [45,46][45][46]. This apo complex is able to bridge a [2Fe-2S]2+ cluster between the two proteins being coordinated, on the GLRX5 side, by the conserved Cys67 and by the cysteine of a GLRX5-bound glutathione (GSH) molecule, and on the BOLA3 side by the conserved Cys59 and His96. This holo-complex has been shown in vitro to function in [2Fe-2S] cluster trafficking in the mitochondrial iron–sulfur protein biogenesis. The [2Fe-2S]2+ BOLA3-GLRX5 complex was shown indeed to transfer the cluster to both apo human ferredoxins FDX1 and FDX2 with rate constants comparable to other cluster donors to FDX proteins [20], as well as to transfer its cluster to apo NFU1 to form a [4Fe-4S]2+ NFU1 dimer [44]. However, considering that the yeast homologue of human BOLA3 was shown not to be required for the maturation of mitochondrial [2Fe-2S] proteins [46], the cluster transfer to FDXs is very likely not physiologically relevant. On the other hand, cluster transfer and assembly from [2Fe-2S]2+ GLRX5-BOLA3 to NFU1 was proposed to be alternative to the pathway involving the [4Fe-4S] cluster transfer from the ISCA1-ISCA2 complex to NFU1, being exclusively activated under oxidative cellular conditions [44] (Figure 1).3. Mutations on Components Maturing ISC Proteins Cause Severe Congenital Diseases
Mutations in GLRX5, which connects the first step of the ISC machinery to its last step, as well as mutations in the accessory proteins, which are involved in the late-acting step of the ISC machinery devoted to mitochondrial [4Fe-4S]-binding protein biogenesis, cause different Fe-S cluster-related diseases, such as sideroblastic anemia, muscle myopathy, multiple mitochondrial disfunction syndromes 1 to 5, and complex I deficiency [23,30,47,48,49][23][30][47][48][49]. All of the aforementioned diseases are associated with severe, often lethal outcomes due to defects in mitochondrial [4Fe-4S] proteins, documenting the importance of these late-acting accessory proteins for cell viability. Here, below, wresearchers report a description of all pathogenic mutations identified up to now in GLRX5 and in the late-acting accessory proteins, and weresearchers describe, from a structural point of view, the effects that these mutations can have on the mutated proteins and on impairing their protein–protein interaction networks.3.1. Structural Aspects of Pathogenic Missense Mutations in GLRX5, a Protein Involved in a Rare Form of Congenital Sideroblastic Anemia
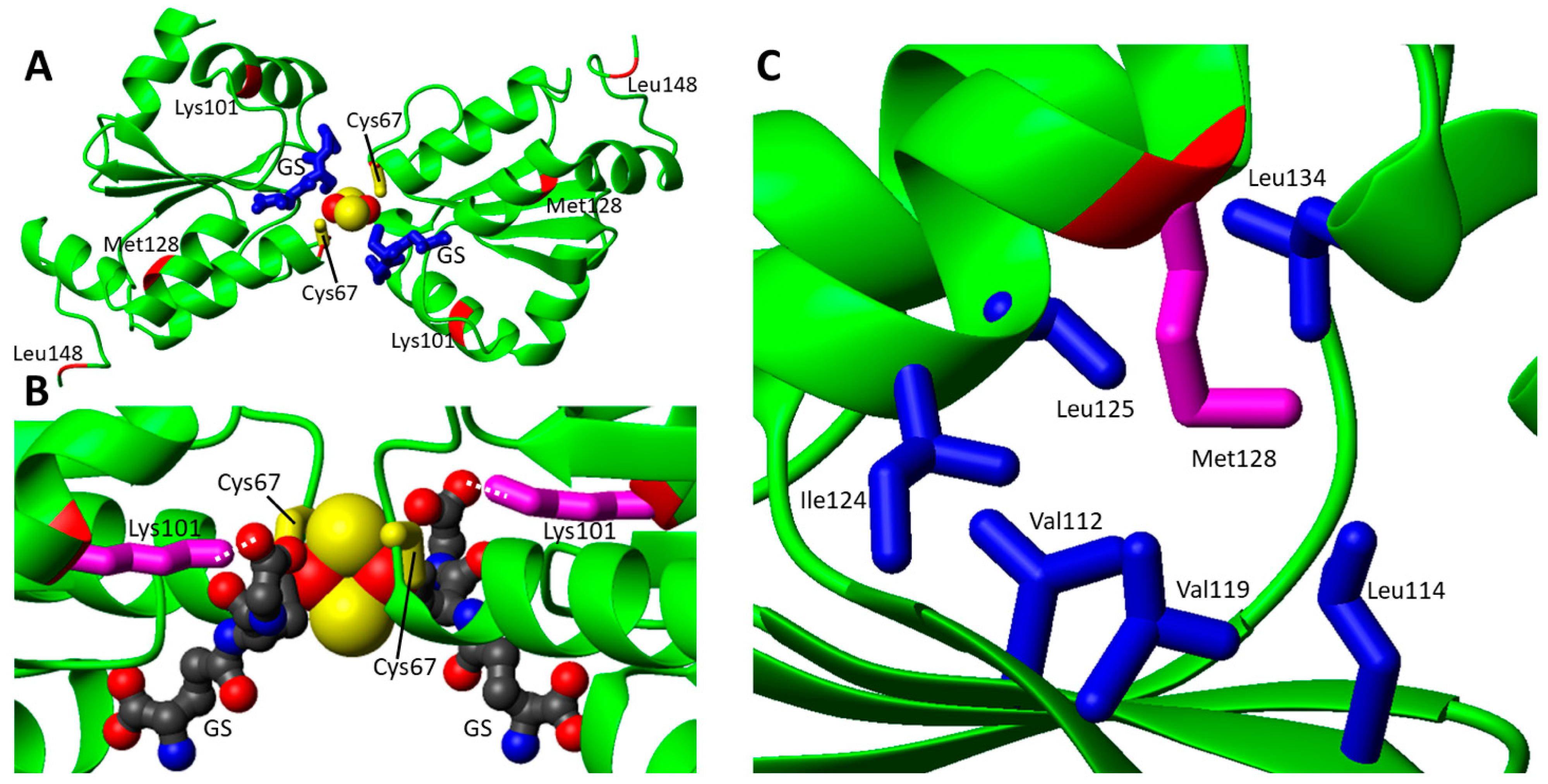
Mutations in GLXR5 gene have been associated so far with two different phenotypes, i.e., the variant nonketotic hyperglycinemia (NKH, MIM# 605899) [50] and the congenital sideroblastic anemia (SIDBA3, MIM#616860) [51]. NKH was associated with the non-sense p.K51del variant of GLRX5 [50]. On the contrary, missense mutations were found in patients affected by SIDBA3. Congenital sideroblastic anemia comprises a heterogeneous group of genetic disorders, characterized by reduced heme synthesis, mitochondrial iron overload and the presence of ringed sideroblasts [24]. In total, four pathogenic missense mutations (two pairs of heterologous pathogenic missense mutations) were identified in GLRX5 gene (Table 1). Two heterozygous missense mutations were found in the GLRX5 gene of a Chinese patient showing SIDBA3: Lys101Gln and Leu148Ser substitution in the GLRX5 protein [51]. Lys101 in GLRX5 protein is highly conserved from yeast to humans [51], whereas Leu148 is less conserved. Decreased holo IRP1 protein levels were observed in peripheral blood mononuclear cells (PBMCs), and, consequently, decreased activity of cytosolic aconitase was evident in PBMCs, together with increased expression of Transferrin receptor 1 (TfR1) protein and reduced expression of H-ferritin protein. Decreased ferrochelatase (FECH) level suggested mitochondrial Fe-S biogenesis impairment in PBMCs of the patient [51]. Daher et al. identified in a 14-year-old girl with SIDBA3, featuring heterozygous mutations in the GLRX5 gene (Cys69Tyr and Met128Lys, Table 1). Functional studies of these variants were not performed, but studies in patient lymphoblastoid cells showed decreased activity in several Fe-S containing enzymes, including mitochondrial respiratory chain complexes I and mitochondrial aconitase (mACO), and in heme-containing enzymes, such as respiratory chain Complex IV [52].| Gene/ Protein |
Missense Mutation | Predicted Protein Mutations | Associated Disease | References |
|---|---|---|---|---|
| GLRX5 | c.301A > C; c.443T > C | p.Lys101Gln; p.Leu148Ser | Nonsindromic sideroblastic anemia 3 | [51] |
| c.200G > A; c.383T > A | p.Cys67Tyr; p.Met128Lys | Nonsindromic sideroblastic anemia 3 | [52] | |
| NFU1 | c.545G > A; c.545G > A | p.Arg182Gln; p.Arg182Gln | MMDS1 | [43] |
| c.622G > T; c.622G > T | p.Gly208Cys; p.Gly208Cys | MMDS1 | [55,56][53][54] | |
| c.565G > A; c.565G > A | p.Gly189Arg; p.Gly189Arg | MMDS1 | [57][55] | |
| c.179G > T; c.179G > T | p.Phe60Cys; p.Phe60Cys | MMDS1 | [58][56] | |
| c.545G > A; c.622G > T | p.Arg182Gln; p.Gly208Cys | MMDS1 | [59][57] | |
| c.544C > T; c.622G > T | p.Arg182Trp; p.Gly208Cys | MMDS1 | [60][58] | |
| c.629G > T; c.622G > T | p.Cys210Phe; p.Gly208Cys | MMDS1 | [58][56] | |
| c.565G > A; c.622G> T | p.Gly189Arg; p.Gly208Cys | MMDS1 | [58,61][56][59] | |
| c.565G > A; c.629G > T | p.Gly189Arg; p.Cys210Phe | MMDS1 | [62][60] | |
| c.565G > A; c.568G > A | p.Gly189Arg; p.Gly190Arg | MMDS1 | [63][61] | |
| c.62G > C; c.622G > T | p.Arg21Pro; p.Gly208Cys | MMDS1 | [63][61] | |
| c.299C > G; c.398T > C | p.Ala100Gly; p.Leu133Pro | MMDS1 | [64][62] | |
| c.721G > T; c.303_369del | p.Val241Phe; ? | MMDS1 | [65][63] | |
| BOLA3 | c.200T > A; c.200T > A | p.Ile67Asn; p.Ile67Asn | MMDS2 | [66,67][64][65] |
| c.287A > G; c.287A > G | p.His96Arg; p.His96Arg | MMDS2 | [68,69,70][66][67][68] | |
| c.295C > T; c.295C > T | p.Arg99Trp; p.Arg99Trp | MMDS2 | [71][69] | |
| c.176G > A; c.136C > T | p.Cys59Tyr; p.Arg46 *a | MMDS2 | [72][70] | |
| IBA57 | c.706C > T; c.706C > T | p.Pro236Ser; p.Pro236Ser | MMDS3 | [73][71] |
| c.941A > C; c.941A > C | p.Gln314Pro; p.Gln314Pro | MMDS3 | [74][72] | |
| c.286T > C; c.188G > A | p.Tyr96His; p.Gly63Asp | MMDS3 | [75,76][73][74] | |
| c.316A > G; c.286T > C | p.Thr106Ala; p.Tyr96His | MMDS3 | [75,76][73][74] | |
| c.738C > G; c.316A > G | p.Asn246Lys; p.Thr106Ala | MMDS3 | [58][56] | |
| c.757G > C; c.316A > G | p.Val253Leu; p.Thr106Ala | MMDS3 | [58][56] | |
| c.335T > G; p.437G > C | p.Leu112Trp; p.Arg146Pro | MMDS3 | [58][56] | |
| c.335T > C; c.588dup | p.Leu112Ser; p.Arg197Alafs | MMDS3 | [77][75] | |
| c.386A > T; c.731A > C | p.Asp129Val; p.Glu244Ala | MMDS3 | [78][76] | |
| c.436C > T; c.436C > T | p.Arg146Trp; p.Arg146Trp | MMDS3 | [79][77] | |
| c.586T > G;c.686C > T | p.Trp196Gly; p.Pro229Leu | MMDS3 | [73][71] | |
| c.656 > G; c.706C > T | p.Tyr219Cys; p.Pro236Ser | MMDS3 | [71][69] | |
| c.701A > G; c.782T > C | p.Asp234Gly; p.Ile261Thr | MMDS3 | [75,76][73][74] | |
| c.738C > G; c.802C > T | p.Asn246Lys; p.Arg268Cys | MMDS3 | [71][69] | |
| c.286T > C; c.754G > T | p.Tyr96His; p.Gly252Cys | MMDS3 | [75,76][73][74] | |
| c.323A > C; c.150C > A | p.Tyr108Ser; pCys50 *a | MMDS3 | [78][76] | |
| c.87insGCCCAAGGTGC; c.313C > T | p.Arg30Alafs; p.Arg105Trp | MMDS3 | [73][71] | |
| c.236C > T; c.307C > T | p.Pro79Leu; p.Gln103 *a | MMDS3 | [76][74] | |
| c.580A > G; c.286T > C | p.Met194Val; p.Tyr96His | MMDS3 | [80][78] | |
| ISCA2 | c.154C > T; c.154C > T | p.Leu52Phe; p.Leu52Phe | MMDS4 | [58][56] |
| c.313A > G; c.313A > G | p.Arg105Gly; p.Arg105Gly | MMDS4 | [58][56] | |
| c.G229 > A; c.G229 > A | p.Gly77Ser; p.Gly77Ser | MMDS4 | [81,82,83][79][80][81] | |
| c.355G > A; c.355G > A | p.Ala119Thr; p.Ala119Thr | MMDS4 | [84][82] | |
| c.5C > A; c.413C > G | p.Ala2Asp; p.Pro138Arg | MMDS4 | [85][83] | |
| c.295delT; c.334A > G | p.Phe99Leufs*18; b p.Ser112Gly | [86][84] | ||
| ISCA1 | c.259G > A; c.259G > A | p.Glu87Lys; p.Glu87Lys | MMDS5 | [87,88][85][86] |
| c.29T > G; c.29T > G | p.Val10Gly; p.Val10Gly | MMDS5 | [89][87] | |
| c.302A > G; c.302A > G | p.Tyr101Cys; p.Tyr101Cys | MMDS5 | [90][88] | |
| IND1 | c.815-27T > C; c.G166 > A | p.Asp273Glnfs*31; b p.Gly56Arg | Complex I deficiency | [91,92,93][89][90][91] |
| c.313G > T; c.166G > A; c.815-27T > C | p.Asp105Tyr; p.Gly56Arg; p.Asp273Glnfs*31 b | Complex I deficiency | [91,92,93][89][90][91] | |
| c.579A > C; c.G166 > A | p.Leu193Phe; p.Gly56Arg | Complex I deficiency | [91,92,93][89][90][91] | |
| c.311T > C; c.815-27T > C | p.Leu104Pro; p.Asp273Glnfs*31 b | Complex I deficiency | [94][92] | |
| c.815-27T > C; c.545T > C | p.Val182Ala; p.Val182Ala | Complex I deficiency | [94][92] | |
| FDX2 | c.1A > T; c.1A > T | p.Met1Leu; p.Met1Leu | MEOAL | [95][93] |
| c.431C > T; c.431C > T | p.Pro144Leu; p.Pro144Leu | MEOAL | [96][94] |
3.2. Pathogenic Missense Mutations in the Late Acting Accessory Proteins of the ISC Machinery
Multiple Mitochondrial Dysfunction Syndromes (MMDS) types 1 to 5 are a group of rare but severe autosomal recessive diseases, caused by variants in the genes encoding for NFU1, BOLA3, IBA57, ISCA2 and ISCA1 proteins, respectively. The hallmark of these diseases is a decreased energy metabolism, which results in defects in neurologic development, muscle weakness, lactic acidosis, respiratory failure. Generally, the pathology manifests in early infancy and results in early death [23]. All of the MMDSs 1–5 have the main impact on enzymes whose functions rely on the presence of bound [4Fe-4S] clusters, such as LIAS and respiratory complexes I and II. In addition to MMDS 1 to 5, two other genetic diseases are linked to genes of the late-acting step of the ISC machinery. The first is called Complex I deficiency, and is associated with mutations in the mitochondrial P-loop NTPase IND1, that is directly responsible for the maturation of the [4Fe-4S] cluster-containing subunits of respiratory chain complex I. A second is called Episodic mitochondrial myopathy with or without optic atrophy and reversible leukoencephalopathy (MEOAL), which has been linked to deficiency of FDX2. FDX2 is a protein that binds a [2Fe-2S] cluster that provides electrons to assemble the [4Fe-4S] cluster and is thus responsible for the maturation of all mitochondrial [4Fe-4S] target proteins.3.2.1. Structural Aspects of Pathogenic Missense Mutations of NFU1 Involved in MMDS1
Bi-allelic pathogenic variants in the NFU1 gene cause MMDS type 1 (MMDS1, MIM#605711), clinically characterized by severe encephalopathy that manifests in the first year of life, with evidences of decreased energetic metabolism. Eleven missense (Table 1) and six non-sense disease-causing variants in NFU1 have been identified to date in patients affected by MMDS1. The metabolic profiles of these patients are characterized by a decreased activity of several mitochondrial proteins, such as pyruvate dehydrogenase (PDH), Glycine Cleavage System (GCS) and respiratory chain complexes. PDH and GCS decreased activity was due to lipoic acid synthesis impairment. The structural characterization of apo NFU1 showed that the apo form is monomeric in solution and adopts a dumbbell-shaped structure with well-structured N- and C-domains connected by a linker [54][96]. It has been also shown that NFU1 binds a [4Fe-4S]2+ cluster, which induces the formation of a homo-dimer through the bridging [4Fe-4S]2+ cluster, coordinated by two cysteines of the conserved CXXC motif of each of the two C-domains [44]. Moreover, the presence of an equilibrium between the [4Fe-4S]2+ NFU1 dimer with the cluster coordinated by the Cys residues of the two CXXC motifs, and a [4Fe-4S]2+ NFU1 dimer where a Cys ligand of the CXXC motif is replaced by a S-donor small molecule ligand, such as GSH or DTT, was observed [44]. In order to analyze the effects of the pathogenic missense mutations on the protein structure, wresearchers have considered the solution NMR structures of the single N- and C-domains of NFU1 (2LTM and 2M5O, respectively), since no structure of the full-length protein is yet available. The missense pathogenic mutations are mainly located on the C-domain of NFU1 in the surrounding of the cluster binding CXXC motif (7 out of 11 mutations, Figure 3A).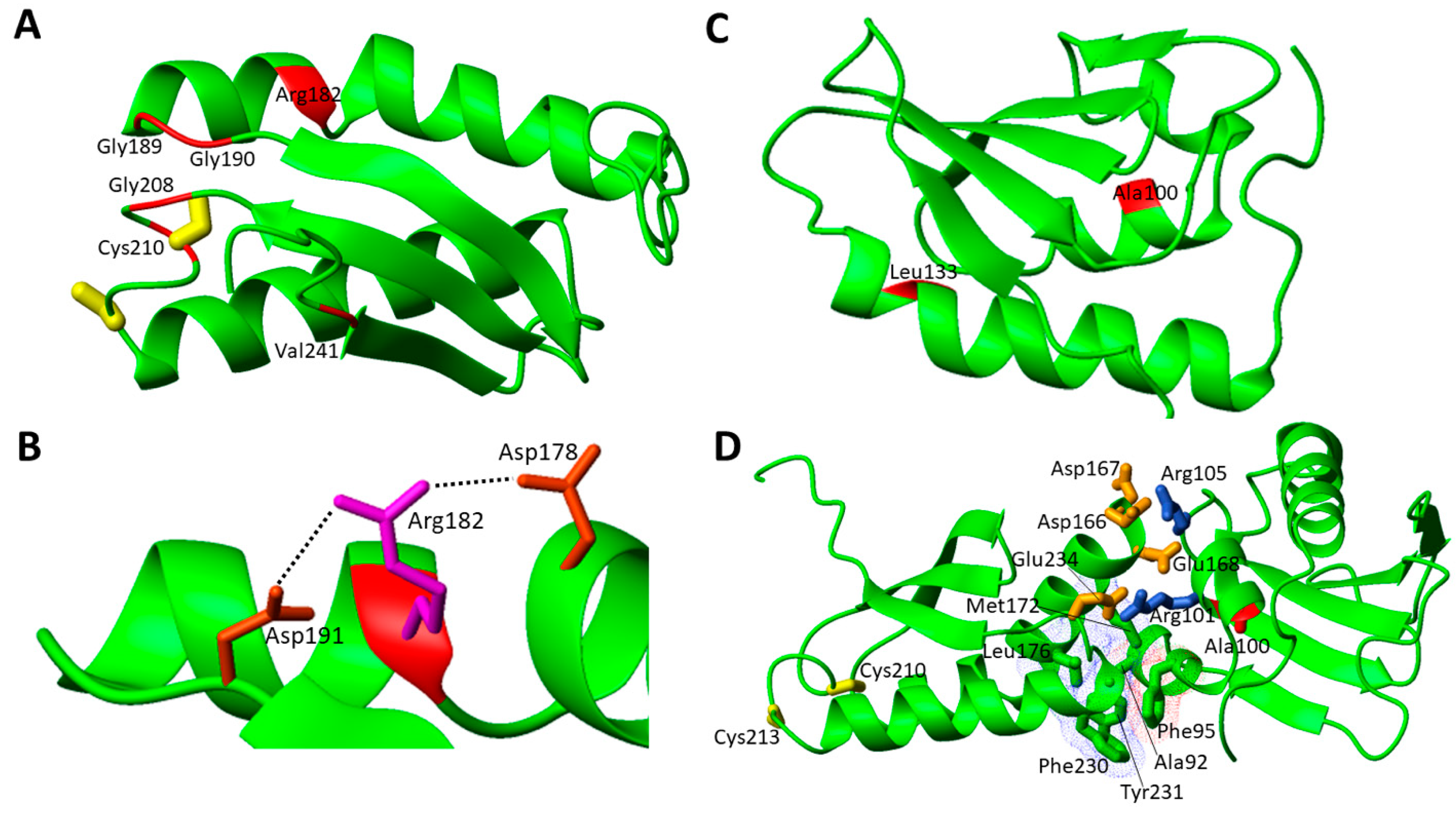
References
- Johannsen, D.L.; Ravussin, E. The Role of Mitochondria in Health and Disease. Curr. Opin. Pharmacol. 2009, 9, 780–786.
- Friedman, J.R.; Nunnari, J. Mitochondrial Form and Function. Nature 2014, 505, 335–343.
- Lill, R. Function and Biogenesis of Iron-Sulphur Proteins. Nature 2009, 460, 831–838.
- Rouault, T.A. The Indispensable Role of Mammalian Iron Sulfur Proteins in Function and Regulation of Multiple Diverse Metabolic Pathways. Biometals 2019, 32, 343–353.
- Beinert, H. Iron-Sulfur Proteins: Ancient Structures, Still Full of Surprises. J. Biol. Inorg. Chem. 2000, 5, 2–15.
- Ciofi-Baffoni, S.; Nasta, V.; Banci, L. Protein Networks in the Maturation of Human Iron–Sulfur Proteins. Metallomics 2018, 10, 49–72.
- Lill, R.; Dutkiewicz, R.; Freibert, S.A.; Heidenreich, T.; Mascarenhas, J.; Netz, D.J.; Paul, V.D.; Pierik, A.J.; Richter, N.; Stümpfig, M.; et al. The Role of Mitochondria and the CIA Machinery in the Maturation of Cytosolic and Nuclear Iron-Sulfur Proteins. Eur. J. Cell Biol. 2015, 94, 280–291.
- Braymer, J.J.; Lill, R. Iron-Sulfur Cluster Biogenesis and Trafficking in Mitochondria. J. Biol. Chem. 2017, 292, 12754–12763.
- Maio, N.; Jain, A.; Rouault, T.A. Mammalian Iron-Sulfur Cluster Biogenesis: Recent Insights into the Roles of Frataxin, Acyl Carrier Protein and ATPase-Mediated Transfer to Recipient Proteins. Curr. Opin. Chem. Biol. 2020, 55, 34–44.
- Boniecki, M.T.; Freibert, S.A.; Mühlenhoff, U.; Lill, R.; Cygler, M. Structure and Functional Dynamics of the Mitochondrial Fe/S Cluster Synthesis Complex. Nat. Commun. 2017, 8, 1287.
- Cory, S.A.; Van Vranken, J.G.; Brignole, E.J.; Patra, S.; Winge, D.R.; Drennan, C.L.; Rutter, J.; Barondeau, D.P. Structure of Human Fe-S Assembly Subcomplex Reveals Unexpected Cysteine Desulfurase Architecture and Acyl-ACP-ISD11 Interactions. Proc. Natl. Acad. Sci. USA 2017, 114, E5325–E5334.
- Patra, S.; Barondeau, D.P. Mechanism of Activation of the Human Cysteine Desulfurase Complex by Frataxin. Proc. Natl. Acad. Sci. USA 2019, 116, 19421–19430.
- Van Vranken, J.G.; Jeong, M.-Y.; Wei, P.; Chen, Y.-C.; Gygi, S.P.; Winge, D.R.; Rutter, J. The Mitochondrial Acyl Carrier Protein (ACP) Coordinates Mitochondrial Fatty Acid Synthesis with Iron Sulfur Cluster Biogenesis. Elife 2016, 5, e17828.
- Adam, A.C.; Bornhövd, C.; Prokisch, H.; Neupert, W.; Hell, K. The Nfs1 Interacting Protein Isd11 Has an Essential Role in Fe/S Cluster Biogenesis in Mitochondria. EMBO J. 2006, 25, 174–183.
- Wiedemann, N.; Urzica, E.; Guiard, B.; Müller, H.; Lohaus, C.; Meyer, H.E.; Ryan, M.T.; Meisinger, C.; Mühlenhoff, U.; Lill, R.; et al. Essential Role of Isd11 in Mitochondrial Iron-Sulfur Cluster Synthesis on Isu Scaffold Proteins. EMBO J. 2006, 25, 184–195.
- Sheftel, A.D.; Stehling, O.; Pierik, A.J.; Elsässer, H.-P.; Mühlenhoff, U.; Webert, H.; Hobler, A.; Hannemann, F.; Bernhardt, R.; Lill, R. Humans Possess Two Mitochondrial Ferredoxins, Fdx1 and Fdx2, with Distinct Roles in Steroidogenesis, Heme, and Fe/S Cluster Biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 11775–11780.
- Cai, K.; Tonelli, M.; Frederick, R.O.; Markley, J.L. Human Mitochondrial Ferredoxin 1 (FDX1) and Ferredoxin 2 (FDX2) Both Bind Cysteine Desulfurase and Donate Electrons for Iron-Sulfur Cluster Biosynthesis. Biochemistry 2017, 56, 487–499.
- Gervason, S.; Larkem, D.; Mansour, A.B.; Botzanowski, T.; Müller, C.S.; Pecqueur, L.; Le Pavec, G.; Delaunay-Moisan, A.; Brun, O.; Agramunt, J.; et al. Physiologically Relevant Reconstitution of Iron-Sulfur Cluster Biosynthesis Uncovers Persulfide-Processing Functions of Ferredoxin-2 and Frataxin. Nat. Commun. 2019, 10, 3566.
- Dutkiewicz, R.; Nowak, M. Molecular Chaperones Involved in Mitochondrial Iron–Sulfur Protein Biogenesis. J. Biol. Inorg. Chem. 2018, 23, 569–579.
- Sen, S.; Rao, B.; Wachnowsky, C.; Cowan, J.A. Cluster Exchange Reactivity of Cluster-Bridged Complexes of BOLA3 with Monothiol Glutaredoxins. Metallomics 2018, 10, 1282–1290.
- Brancaccio, D.; Gallo, A.; Mikolajczyk, M.; Zovo, K.; Palumaa, P.; Novellino, E.; Piccioli, M.; Ciofi-Baffoni, S.; Banci, L. Formation of Clusters in the Mitochondrial Iron-Sulfur Cluster Assembly Machinery. J. Am. Chem. Soc. 2014, 136, 16240–16250.
- Banci, L.; Brancaccio, D.; Ciofi-Baffoni, S.; Del Conte, R.; Gadepalli, R.; Mikolajczyk, M.; Neri, S.; Piccioli, M.; Winkelmann, J. Cluster Transfer in Iron-Sulfur Protein Biogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 6203–6208.
- Lebigot, E.; Schiff, M.; Golinelli-Cohen, M.-P. A Review of Multiple Mitochondrial Dysfunction Syndromes, Syndromes Associated with Defective Fe-S Protein Maturation. Biomedicines 2021, 9, 989.
- Camaschella, C. Recent Advances in the Understanding of Inherited Sideroblastic Anaemia. Br. J. Haematol. 2008, 143, 27–38.
- Jain, A.; Singh, A.; Maio, N.; Rouault, T.A. Assembly of the Cluster of NFU1 Requires the Coordinated Donation of Two Clusters from the Scaffold Proteins, ISCU2 and ISCA1. Hum. Mol. Genet. 2020, 29, 3165–3182.
- Brancaccio, D.; Gallo, A.; Piccioli, M.; Novellino, E.; Ciofi-Baffoni, S.; Banci, L. Cluster Assembly in Mitochondria and Its Impairment by Copper. J. Am. Chem. Soc. 2017, 139, 719–730.
- Weiler, B.D.; Brück, M.-C.; Kothe, I.; Bill, E.; Lill, R.; Mühlenhoff, U. Mitochondrial Protein Assembly Involves Reductive Cluster Fusion on ISCA1-ISCA2 by Electron Flow from Ferredoxin FDX2. Proc. Natl. Acad. Sci. USA 2020, 117, 20555–20565.
- Gourdoupis, S.; Nasta, V.; Calderone, V.; Ciofi-Baffoni, S.; Banci, L. IBA57 Recruits ISCA2 to Form a Cluster-Mediated Complex. J. Am. Chem. Soc. 2018, 140, 14401–14412.
- Nasta, V.; Da Vela, S.; Gourdoupis, S.; Ciofi-Baffoni, S.; Svergun, D.I.; Banci, L. Structural Properties of ISCA2-IBA57: A Complex of the Mitochondrial Iron-Sulfur Cluster Assembly Machinery. Sci. Rep. 2019, 9, 18986.
- Stehling, O.; Wilbrecht, C.; Lill, R. Mitochondrial Iron-Sulfur Protein Biogenesis and Human Disease. Biochimie 2014, 100, 61–77.
- Maio, N.; Rouault, T.A. Iron-Sulfur Cluster Biogenesis in Mammalian Cells: New Insights into the Molecular Mechanisms of Cluster Delivery. Biochim. Biophys. Acta 2015, 1853, 1493–1512.
- Saudino, G.; Ciofi-Baffoni, S.; Banci, L. Protein-Interaction Affinity Gradient Drives Cluster Insertion in Human Lipoyl Synthase. J. Am. Chem. Soc. 2022, 144, 5713–5717.
- Suraci, D.; Saudino, G.; Nasta, V.; Ciofi-Baffoni, S.; Banci, L. ISCA1 Orchestrates ISCA2 and NFU1 in the Maturation of Human Mitochondrial Proteins. J. Mol. Biol. 2021, 433, 166924.
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-Adenosylmethionine Enzymes. Chem. Rev. 2014, 114, 4229–4317.
- Dowling, D.P.; Vey, J.L.; Croft, A.K.; Drennan, C.L. Structural Diversity in the AdoMet Radical Enzyme Superfamily. Biochim. Biophys. Acta—Proteins Proteom. 2012, 1824, 1178–1195.
- Cicchillo, R.M.; Iwig, D.F.; Jones, A.D.; Nesbitt, N.M.; Baleanu-Gogonea, C.; Souder, M.G.; Tu, L.; Booker, S.J. Lipoyl Synthase Requires Two Equivalents of S-Adenosyl-L-Methionine to Synthesize One Equivalent of Lipoic Acid. Biochemistry 2004, 43, 6378–6386.
- Lanz, N.D.; Booker, S.J. Identification and Function of Auxiliary Iron-Sulfur Clusters in Radical SAM Enzymes. Biochim. Biophys. Acta 2012, 1824, 1196–1212.
- Fontecave, M.; Ollagnier-de-Choudens, S.; Mulliez, E. Biological Radical Sulfur Insertion Reactions. Chem. Rev. 2003, 103, 2149–2166.
- Camponeschi, F.; Muzzioli, R.; Ciofi-Baffoni, S.; Piccioli, M.; Banci, L. Paramagnetic 1H NMR Spectroscopy to Investigate the Catalytic Mechanism of Radical S-Adenosylmethionine Enzymes. J. Mol. Biol. 2019, 431, 4514–4522.
- Hendricks, A.L.; Wachnowsky, C.; Fries, B.; Fidai, I.; Cowan, J.A. Characterization and Reconstitution of Human Lipoyl Synthase (LIAS) Supports ISCA2 and ISCU as Primary Cluster Donors and an Ordered Mechanism of Cluster Assembly. Int. J. Mol. Sci. 2021, 22, 1598.
- Lanz, N.D.; Rectenwald, J.M.; Wang, B.; Kakar, E.S.; Laremore, T.N.; Booker, S.J.; Silakov, A. Characterization of a Radical Intermediate in Lipoyl Cofactor Biosynthesis. J. Am. Chem. Soc. 2015, 137, 13216–13219.
- McLaughlin, M.I.; Lanz, N.D.; Goldman, P.J.; Lee, K.-H.; Booker, S.J.; Drennan, C.L. Crystallographic Snapshots of Sulfur Insertion by Lipoyl Synthase. Proc. Natl. Acad. Sci. USA 2016, 113, 9446–9450.
- Cameron, J.M.; Janer, A.; Levandovskiy, V.; Mackay, N.; Rouault, T.A.; Tong, W.-H.; Ogilvie, I.; Shoubridge, E.A.; Robinson, B.H. Mutations in Iron-Sulfur Cluster Scaffold Genes NFU1 and BOLA3 Cause a Fatal Deficiency of Multiple Respiratory Chain and 2-Oxoacid Dehydrogenase Enzymes. Am. J. Hum. Genet. 2011, 89, 486–495.
- Nasta, V.; Suraci, D.; Gourdoupis, S.; Ciofi-Baffoni, S.; Banci, L. A Pathway for Assembling 2+ Clusters in Mitochondrial Iron-Sulfur Protein Biogenesis. FEBS J. 2020, 287, 2312–2327.
- Nasta, V.; Giachetti, A.; Ciofi-Baffoni, S.; Banci, L. Structural Insights into the Molecular Function of Human BOLA1-GRX5 and BOLA3-GRX5 Complexes. Biochim. Biophys. Acta 2017, 1861, 2119–2131.
- Uzarska, M.A.; Nasta, V.; Weiler, B.D.; Spantgar, F.; Ciofi-Baffoni, S.; Saviello, M.R.; Gonnelli, L.; Mühlenhoff, U.; Banci, L.; Lill, R. Mitochondrial Bol1 and Bol3 Function as Assembly Factors for Specific Iron-Sulfur Proteins. Elife 2016, 5, e16673.
- Maio, N.; Rouault, T.A. Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis. Trends Biochem. Sci. 2020, 45, 411–426.
- Maio, N.; Rouault, T.A. Mammalian Iron Sulfur Cluster Biogenesis and Human Diseases. IUBMB Life 2022, 74, 705–714.
- Wachnowsky, C.; Fidai, I.; Cowan, J.A. Iron–Sulfur Cluster Biosynthesis and Trafficking—Impact on Human Disease Conditions. Metallomics 2018, 10, 9–29.
- Baker, P.R.; Friederich, M.W.; Swanson, M.A.; Shaikh, T.; Bhattacharya, K.; Scharer, G.H.; Aicher, J.; Creadon-Swindell, G.; Geiger, E.; MacLean, K.N.; et al. Variant Non Ketotic Hyperglycinemia Is Caused by Mutations in LIAS, BOLA3 and the Novel Gene GLRX5. Brain 2014, 137, 366–379.
- Liu, G.; Guo, S.; Anderson, G.J.; Camaschella, C.; Han, B.; Nie, G. Heterozygous Missense Mutations in the GLRX5 Gene Cause Sideroblastic Anemia in a Chinese Patient. Blood 2014, 124, 2750–2751.
- Daher, R.; Mansouri, A.; Martelli, A.; Bayart, S.; Manceau, H.; Callebaut, I.; Moulouel, B.; Gouya, L.; Puy, H.; Kannengiesser, C.; et al. GLRX5 Mutations Impair Heme Biosynthetic Enzymes ALA Synthase 2 and Ferrochelatase in Human Congenital Sideroblastic Anemia. Mol. Genet. Metab. 2019, 128, 342–351.
- Navarro-Sastre, A.; Tort, F.; Stehling, O.; Uzarska, M.A.; Arranz, J.A.; Del Toro, M.; Labayru, M.T.; Landa, J.; Font, A.; Garcia-Villoria, J.; et al. A Fatal Mitochondrial Disease Is Associated with Defective NFU1 Function in the Maturation of a Subset of Mitochondrial Fe-S Proteins. Am. J. Hum. Genet. 2011, 89, 656–667.
- Stéphanie, P.; Catherine, B.; Thierry, S.; Jean-Luc, V.; Isabelle, L.; Sara, S.; Christophe, V.; Marie-Cécile, N. “Idiopathic” Pulmonary Arterial Hypertension in Early Infancy: Excluding NFU1 Deficiency. Ann. Pediatr. Cardiol. 2019, 12, 325–328.
- Uzunhan, T.A.; Çakar, N.E.; Seyhan, S.; Aydin, K. A Genetic Mimic of Cerebral Palsy: Homozygous NFU1 Mutation with Marked Intrafamilial Phenotypic Variation. Brain Dev. 2020, 42, 756–761.
- Lebigot, E.; Gaignard, P.; Dorboz, I.; Slama, A.; Rio, M.; de Lonlay, P.; Héron, B.; Sabourdy, F.; Boespflug-Tanguy, O.; Cardoso, A.; et al. Impact of Mutations within the Cluster or the Lipoic Acid Biosynthesis Pathways on Mitochondrial Protein Expression Profiles in Fibroblasts from Patients. Mol. Genet. Metab. 2017, 122, 85–94.
- de Souza, P.V.S.; Bortholin, T.; Burlin, S.; Naylor, F.G.M.; de Rezende Pinto, W.B.V.; Oliveira, A.S.B. NFU1 -Related Disorders as Key Differential Diagnosis of Cavitating Leukoencephalopathy. J. Pediatr. Genet. 2018, 7, 40–42.
- Birjiniuk, A.; Glinton, K.E.; Villafranco, N.; Boyer, S.; Laufman, J.; Mizerik, E.; Scott, D.; Elsea, S.H.; Galambos, C.; Varghese, N.P.; et al. Multiple Mitochondrial Dysfunctions Syndrome 1: An Unusual Cause of Developmental Pulmonary Hypertension. Am. J. Med. Genet. Part A 2020, 182, 755–761.
- Nizon, M.; Boutron, A.; Boddaert, N.; Slama, A.; Delpech, H.; Sardet, C.; Brassier, A.; Habarou, F.; Delahodde, A.; Correia, I.; et al. Leukoencephalopathy with Cysts and Hyperglycinemia May Result from NFU1 Deficiency. Mitochondrion 2014, 15, 59–64.
- Invernizzi, F.; Ardissone, A.; Lamantea, E.; Garavaglia, B.; Zeviani, M.; Farina, L.; Ghezzi, D.; Moroni, I. Cavitating Leukoencephalopathy with Multiple Mitochondrial Dysfunction Syndrome and NFU1 Mutations. Front. Genet. 2014, 5, 412.
- Ahting, U.; Mayr, J.A.; Vanlander, A.V.; Hardy, S.A.; Santra, S.; Makowski, C.; Alston, C.L.; Zimmermann, F.A.; Abela, L.; Plecko, B.; et al. Clinical, Biochemical, and Genetic Spectrum of Seven Patients with NFU1 Deficiency. Front. Genet. 2015, 6, 123.
- Ames, E.G.; Neville, K.L.; McNamara, N.A.; Keegan, C.E.; Elsea, S.H. Clinical Reasoning: A 12-Month-Old Child with Hypotonia and Developmental Delays. Neurology 2020, 95, 184–187.
- Jin, D.; Yu, T.; Zhang, L.; Wang, T.; Hu, J.; Wang, Y.; Yang, X.-A. Novel NFU1 Variants Induced MMDS Behaved as Special Leukodystrophy in Chinese Sufferers. J. Mol. Neurosci. 2017, 62, 255–261.
- Haack, T.B.; Rolinski, B.; Haberberger, B.; Zimmermann, F.; Schum, J.; Strecker, V.; Graf, E.; Athing, U.; Hoppen, T.; Wittig, I.; et al. Homozygous Missense Mutation in BOLA3 Causes Multiple Mitochondrial Dysfunctions Syndrome in Two Siblings. J. Inherit. Metab. Dis. 2013, 36, 55–62.
- Nikam, R.M.; Gripp, K.W.; Choudhary, A.K.; Kandula, V. Imaging Phenotype of Multiple Mitochondrial Dysfunction Syndrome 2, a Rare BOLA3-Associated Leukodystrophy. Am. J. Med. Genet. A 2018, 176, 2787–2790.
- Kohda, M.; Tokuzawa, Y.; Kishita, Y.; Nyuzuki, H.; Moriyama, Y.; Mizuno, Y.; Hirata, T.; Yatsuka, Y.; Yamashita-Sugahara, Y.; Nakachi, Y.; et al. A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet. 2016, 12, e1005679.
- Nishioka, M.; Inaba, Y.; Motobayashi, M.; Hara, Y.; Numata, R.; Amano, Y.; Shingu, K.; Yamamoto, Y.; Murayama, K.; Ohtake, A.; et al. An Infant Case of Diffuse Cerebrospinal Lesions and Cardiomyopathy Caused by a BOLA3 Mutation. Brain Dev. 2018, 40, 484–488.
- Imai-Okazaki, A.; Kishita, Y.; Kohda, M.; Mizuno, Y.; Fushimi, T.; Matsunaga, A.; Yatsuka, Y.; Hirata, T.; Harashima, H.; Takeda, A.; et al. Cardiomyopathy in Children with Mitochondrial Disease: Prognosis and Genetic Background. Int. J. Cardiol. 2019, 279, 115–121.
- Bindu, P.S.; Sonam, K.; Chiplunkar, S.; Govindaraj, P.; Nagappa, M.; Vekhande, C.C.; Aravinda, H.R.; Ponmalar, J.J.; Mahadevan, A.; Gayathri, N.; et al. Mitochondrial Leukoencephalopathies: A Border Zone between Acquired and Inherited White Matter Disorders in Children? Mult. Scler. Relat. Disord. 2018, 20, 84–92.
- Stutterd, C.A.; Lake, N.J.; Peters, H.; Lockhart, P.J.; Taft, R.J.; van der Knaap, M.S.; Vanderver, A.; Thorburn, D.R.; Simons, C.; Leventer, R.J. Severe Leukoencephalopathy with Clinical Recovery Caused by Recessive BOLA3 Mutations. JIMD Rep. 2018, 43, 63–70.
- Torraco, A.; Ardissone, A.; Invernizzi, F.; Rizza, T.; Fiermonte, G.; Niceta, M.; Zanetti, N.; Martinelli, D.; Vozza, A.; Verrigni, D.; et al. Novel Mutations in IBA57 Are Associated with Leukodystrophy and Variable Clinical Phenotypes. J. Neurol. 2017, 264, 102–111.
- Ajit Bolar, N.; Vanlander, A.V.; Wilbrecht, C.; Van der Aa, N.; Smet, J.; De Paepe, B.; Vandeweyer, G.; Kooy, F.; Eyskens, F.; De Latter, E.; et al. Mutation of the Iron-Sulfur Cluster Assembly Gene IBA57 Causes Severe Myopathy and Encephalopathy. Hum. Mol. Genet. 2013, 22, 2590–2602.
- Liu, M.; Zhang, J.; Zhang, Z.; Zhou, L.; Jiang, Y.; Wang, J.; Xiao, J.; Wu, Y. Phenotypic Spectrum of Mutations in IBA57, a Candidate Gene for Cavitating Leukoencephalopathy. Clin. Genet. 2018, 93, 235–241.
- Zhang, J.; Liu, M.; Zhang, Z.; Zhou, L.; Kong, W.; Jiang, Y.; Wang, J.; Xiao, J.; Wu, Y. Genotypic Spectrum and Natural History of Cavitating Leukoencephalopathies in Childhood. Pediatr. Neurol. 2019, 94, 38–47.
- Hamanaka, K.; Miyatake, S.; Zerem, A.; Lev, D.; Blumkin, L.; Yokochi, K.; Fujita, A.; Imagawa, E.; Iwama, K.; Nakashima, M.; et al. Expanding the Phenotype of IBA57 Mutations: Related Leukodystrophy Can Remain Asymptomatic. J. Hum. Genet. 2018, 63, 1223–1229.
- Ishiyama, A.; Sakai, C.; Matsushima, Y.; Noguchi, S.; Mitsuhashi, S.; Endo, Y.; Hayashi, Y.K.; Saito, Y.; Nakagawa, E.; Komaki, H.; et al. IBA57 Mutations Abrogate Iron-Sulfur Cluster Assembly Leading to Cavitating Leukoencephalopathy. Neurol. Genet. 2017, 3, e184.
- Debray, F.-G.; Stümpfig, C.; Vanlander, A.V.; Dideberg, V.; Josse, C.; Caberg, J.-H.; Boemer, F.; Bours, V.; Stevens, R.; Seneca, S.; et al. Mutation of the Iron-Sulfur Cluster Assembly Gene IBA57 Causes Fatal Infantile Leukodystrophy. J. Inherit. Metab. Dis. 2015, 38, 1147–1153.
- Zhan, F.; Liu, X.; Ni, R.; Liu, T.; Cao, Y.; Wu, J.; Tian, W.; Luan, X.; Cao, L. Novel IBA57 Mutations in Two Chinese Patients and Literature Review of Multiple Mitochondrial Dysfunction Syndrome. Metab. Brain Dis. 2022, 37, 311–317.
- Al-Hassnan, Z.N.; Al-Dosary, M.; Alfadhel, M.; Faqeih, E.A.; Alsagob, M.; Kenana, R.; Almass, R.; Al-Harazi, O.S.; Al-Hindi, H.; Malibari, O.I.; et al. ISCA2 Mutation Causes Infantile Neurodegenerative Mitochondrial Disorder. J. Med. Genet. 2015, 52, 186–194.
- Alaimo, J.T.; Besse, A.; Alston, C.L.; Pang, K.; Appadurai, V.; Samanta, M.; Smpokou, P.; McFarland, R.; Taylor, R.W.; Bonnen, P.E. Loss-of-function Mutations in ISCA2 Disrupt 4Fe–4S Cluster Machinery and Cause a Fatal Leukodystrophy with Hyperglycinemia and MtDNA Depletion. Hum. Mutat. 2018, 39, 537–549.
- Alfadhel, M.; Nashabat, M.; Alrifai, M.T.; Alshaalan, H.; Al Mutairi, F.; Al-Shahrani, S.A.; Plecko, B.; Almass, R.; Alsagob, M.; Almutairi, F.B.; et al. Further Delineation of the Phenotypic Spectrum of ISCA2 Defect: A Report of Ten New Cases. Eur. J. Paediatr. Neurol. 2018, 22, 46–55.
- Eidi, M.; Garshasbi, M. A Novel ISCA2 Variant Responsible for an Early-Onset Neurodegenerative Mitochondrial Disorder: A Case Report of Multiple Mitochondrial Dysfunctions Syndrome 4. BMC Neurol. 2019, 19, 153.
- Hartman, T.G.; Yosovich, K.; Michaeli, H.G.; Blumkin, L.; Ben-Sira, L.; Lev, D.; Lerman-Sagie, T.; Zerem, A. Expanding the Genotype-Phenotype Spectrum of ISCA2-Related Multiple Mitochondrial Dysfunction Syndrome-Cavitating Leukoencephalopathy and Prolonged Survival. Neurogenetics 2020, 21, 243–249.
- Toldo, I.; Nosadini, M.; Boscardin, C.; Talenti, G.; Manara, R.; Lamantea, E.; Legati, A.; Ghezzi, D.; Perilongo, G.; Sartori, S. Neonatal Mitochondrial Leukoencephalopathy with Brain and Spinal Involvement and High Lactate: Expanding the Phenotype of ISCA2 Gene Mutations. Metab. Brain Dis. 2018, 33, 805–812.
- Shukla, A.; Hebbar, M.; Srivastava, A.; Kadavigere, R.; Upadhyai, P.; Kanthi, A.; Brandau, O.; Bielas, S.; Girisha, K.M. Homozygous p.(Glu87Lys) Variant in ISCA1 Is Associated with a Multiple Mitochondrial Dysfunctions Syndrome. J. Hum. Genet. 2017, 62, 723–727.
- Shukla, A.; Kaur, P.; Girisha, K.M. Report of the Third Family with Multiple Mitochondrial Dysfunctions Syndrome 5 Caused by the Founder Variant p.(Glu87Lys) in ISCA1. J. Pediatr. Genet. 2018, 7, 130–133.
- Torraco, A.; Stehling, O.; Stümpfig, C.; Rösser, R.; De Rasmo, D.; Fiermonte, G.; Verrigni, D.; Rizza, T.; Vozza, A.; Di Nottia, M.; et al. ISCA1 Mutation in a Patient with Infantile-Onset Leukodystrophy Causes Defects in Mitochondrial Proteins. Hum. Mol. Genet. 2018, 27, 2739–2754.
- Lebigot, E.; Hully, M.; Amazit, L.; Gaignard, P.; Michel, T.; Rio, M.; Lombès, M.; Thérond, P.; Boutron, A.; Golinelli-Cohen, M.P. Expanding the Phenotype of Mitochondrial Disease: Novel Pathogenic Variant in ISCA1 Leading to Instability of the Iron-Sulfur Cluster in the Protein. Mitochondrion 2020, 52, 75–82.
- Kevelam, S.H.; Rodenburg, R.J.; Wolf, N.I.; Ferreira, P.; Lunsing, R.J.; Nijtmans, L.G.; Mitchell, A.; Arroyo, H.A.; Rating, D.; Vanderver, A.; et al. NUBPL Mutations in Patients with Complex I Deficiency and a Distinct MRI Pattern. Neurology 2013, 80, 1577–1583.
- Calvo, S.E.; Tucker, E.J.; Compton, A.G.; Kirby, D.M.; Crawford, G.; Burtt, N.P.; Rivas, M.; Guiducci, C.; Bruno, D.L.; Goldberger, O.A.; et al. High-Throughput, Pooled Sequencing Identifies Mutations in NUBPL and FOXRED1 in Human Complex I Deficiency. Nat. Genet. 2010, 42, 851–858.
- Tucker, E.J.; Mimaki, M.; Compton, A.G.; McKenzie, M.; Ryan, M.T.; Thorburn, D.R. Next-Generation Sequencing in Molecular Diagnosis: NUBPL Mutations Highlight the Challenges of Variant Detection and Interpretation. Hum. Mutat. 2012, 33, 411–418.
- Kimonis, V.; Al Dubaisi, R.; Maclean, A.E.; Hall, K.; Weiss, L.; Stover, A.E.; Schwartz, P.H.; Berg, B.; Cheng, C.; Parikh, S.; et al. NUBPL Mitochondrial Disease: New Patients and Review of the Genetic and Clinical Spectrum. J. Med. Genet. 2021, 58, 314–325.
- Spiegel, R.; Saada, A.; Halvardson, J.; Soiferman, D.; Shaag, A.; Edvardson, S.; Horovitz, Y.; Khayat, M.; Shalev, S.A.; Feuk, L.; et al. Deleterious Mutation in FDX1L Gene Is Associated with a Novel Mitochondrial Muscle Myopathy. Eur. J. Hum. Genet. 2014, 22, 902–906.
- Gurgel-Giannetti, J.; Lynch, D.S.; De Paiva, A.R.B.; Lucato, L.; Yamamoto, G.; Thomsen, C.; Basu, S.; Freua, F.; Giannetti, A.V.; Assis, B.D.R.D.; et al. A Novel Complex Neurological Phenotype Due to a Homozygous Mutation in FDX2. Brain 2018, 141, 2289–2298.
- Johansson, C.; Roos, A.K.; Montano, S.J.; Sengupta, R.; Filippakopoulos, P.; Guo, K.; von Delft, F.; Holmgren, A.; Oppermann, U.; Kavanagh, K.L. The Crystal Structure of Human GLRX5: Iron-Sulfur Cluster Co-Ordination, Tetrameric Assembly and Monomer Activity. Biochem. J. 2011, 433, 303–311.
- Cai, K.; Liu, G.; Frederick, R.O.; Xiao, R.; Montelione, G.T.; Markley, J.L. Structural/Functional Properties of Human NFU1, an Intermediate Carrier in Human Mitochondrial Iron-Sulfur Cluster Biogenesis. Structure 2016, 24, 2080–2091.
- Wachnowsky, C.; Wesley, N.A.; Fidai, I.; Cowan, J.A. Understanding the Molecular Basis of Multiple Mitochondrial Dysfunctions Syndrome 1 (MMDS1)-Impact of a Disease-Causing Gly208Cys Substitution on Structure and Activity of NFU1 in the Fe/S Cluster Biosynthetic Pathway. J. Mol. Biol. 2017, 429, 790–807.
- Tonduti, D.; Dorboz, I.; Imbard, A.; Slama, A.; Boutron, A.; Pichard, S.; Elmaleh, M.; Vallée, L.; Benoist, J.F.; Ogier, H.; et al. New Spastic Paraplegia Phenotype Associated to Mutation of NFU1. Orphanet J. Rare Dis. 2015, 10, 13.
- Wesley, N.A.; Wachnowsky, C.; Fidai, I.; Cowan, J.A. Understanding the Molecular Basis for Multiple Mitochondrial Dysfunctions Syndrome 1 (MMDS1): Impact of a Disease-Causing Gly189Arg Substitution on NFU1. FEBS J. 2017, 284, 3838–3848.
- Tong, W.-H.; Jameson, G.N.L.; Huynh, B.H.; Rouault, T.A. Subcellular Compartmentalization of Human Nfu, an Iron-Sulfur Cluster Scaffold Protein, and Its Ability to Assemble a Cluster. Proc. Natl. Acad. Sci. USA 2003, 100, 9762–9767.
