Salt bridges are interactions, electrostatic combined with hydrogen bonding, between oppositely charged residues, typically carboxylic acid anions and ammonium ions, provided they are close together. For an illustration see Fig. 1. Salt bridges are of particular interest in proteins and other biomolecules. In the present contribution salt bridges are investigated by means of 1H chemical shifts, determination of pKa values and deuterium isotope effect on 15N and 1H chemical shifts. In the latter case model compounds like ammonium ions are also investigated and the use of deuterium isotope effects on chemical shifts are supported by Density Functional Theory (DFT) calculations. The use of isotope effects on chemical shifts enables a distinction between salt bridges observed in the solid state by X-ray diffraction and those actually present in solution.
- deuterium isotope effects on chemical shifts
- salt bridges
- proteins
- theoretical calculations
1. IDefintroductition
Salt bridges may contrilbute to protein stability. They may occur both in the interior and at the surfaceare interactions, electrostatic combined with hydrogen bonding, between oppositely charged residues, typically carboxylic acid anions and ammonium ions, provided they are close together. For an illustration see Fig. 1. Salt bridges could be important for e.g. protein stability.
Salt bridges have traditionally been investigated by measuring changes in pKa values, the mutation approach, X-ray studies of crystalline samples. These techniques are reviewed. [1]
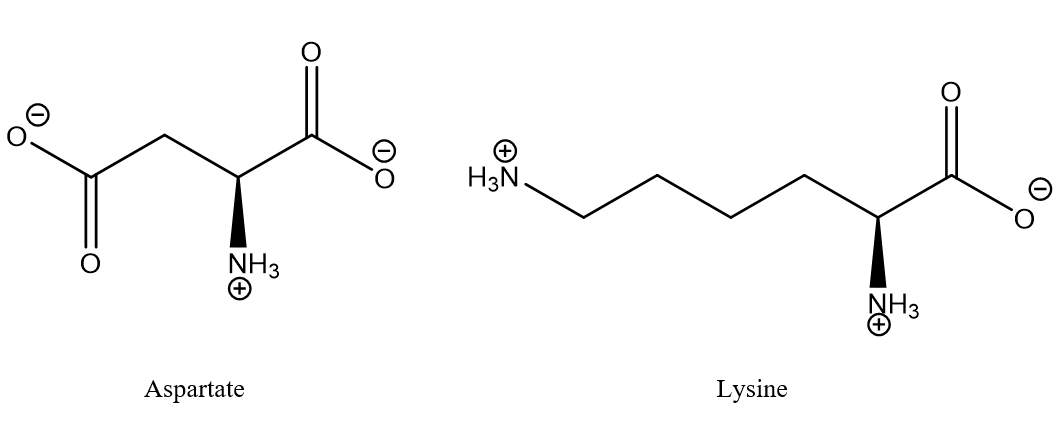
Figure 1. Typical salt bridge between aspartic acid and lysine. The aminoacids may typically be part of different strands of proteins
Salt bridges in the present paper are investigated using 1H NMR chemical shifts, determination of pKa values by NMR and deuterium isotope effects on 15N and 1H chemical shifts. In the latter case model compounds like ammonium salts are investigated and the use of isotope effects on chemical shifts are supported by Density Functional Theory (DFT) calculations (see later). The use of isotope effects on chemical shifts enables a distinction between salt bridges observed in the solid state by X-ray diffraction and those actually present in solution.
2. NMR Titration Studies
A classic case is that of Bovine Pancreatic Trypsin Inhibitor (BPTI) in which the terminals (the C- and the N-terminal) forms a salt bridge in solution, but not in the crystal state. This was demonstrated early on by Wüthrich and co-workers. [2]
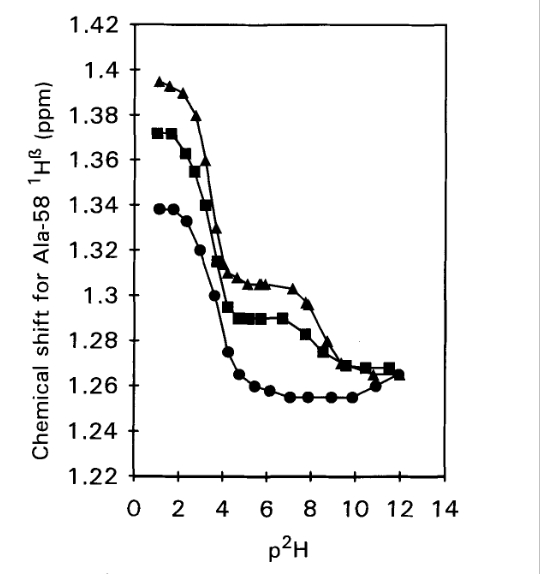
Figure 2. 1H chemical shifts vs p2H for the C-terminal Alanine 58 H-α resonance in WT-BPTI (triangles), 1-BPTI (squares) and 4-BPTI (dots). Samples contained 2 mM protein in 0.3 M KC1 and spectra were recorded at 25°C, 250 MHz. From Ref. [3] with permission from Elsevier.
The idea of determining the pKa values is, that for the acid, a drop in pKa value is expected, if a salt bridge is formed. This is indeed seen in Fig. 2. For the WT-BPTI the value is 3.33, and for 1-BPTI the pKa value is 3.35, whereas for 4-BPTI, not being able to form a salt bridge, it is 3.67. 4-BPTI has four extra amino acids at the N-terminus. [3] This picture is confirmed for 3-58BPTI missing to residues at the N-terminus and not being able to form a salt bridge. In that case the pKa value is 3.72. [4]
The pKa values of e.g. can also be determined by NMR. In that case an increase of the pKa value is expected. For the lysines of protein G B1 domain this was not found in line with the finding (see later), that no salt bridges with lysines exist is this protein in solution. [5]
3. NH Chemical Shifts
The formation of a salt bridge in the complex between dihydrofolate reductase and methothrexate, brodimoprim 4-carboxylate or brodimoprim-4,6-dicarboxylate was established between the ligand and Argine 57 based on the finding of a higher NH chemical shift for those NH protons being part of the salt bridge compared to those not. [6]
4. Use of Isotope Effects on Chemical Shifts to Study Salt Bridges
4.1. Isotope effects on chemical shifts
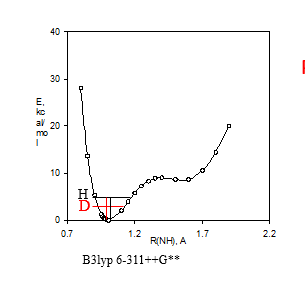
Exchange of a 15NH with a deuterium leads to a change in the 15N chemical shift as illustrated in Fig. 3. Upon deuteriation the vibration sits lower in the potential well. This leads on the average to a shorter ND bond, which as seen in Fig. 3b leads to a change in the 15N nuclear shielding (equal to a smaller chemical shift). The difference between the chemical shift of the deuteriated species minus the protio on is called the isotope effect, nΔ(D)= δX(H)-δX(D). If n is one it is called a one-bond deuterium isotope effect. If X is nitrogen, 1ΔN(D). A negative two bond deuterium isotope effect is observed at the hydrogens, 2ΔH(D). As isotope effects are measured as a difference in chemical shifts in the same solution, they can be measured very accurately. A prerequisite is of course a that separate signals can be observed. For reviews on isotope effects on chemical shifts. [7]
Figure 3. a. Potential energy well. B3lyp 6-311++G** shows the DFT fuctional and basis set used. b. Calculated 15N nuclear shielding surface vs. NH bond length.
4.2. Use of model compounds
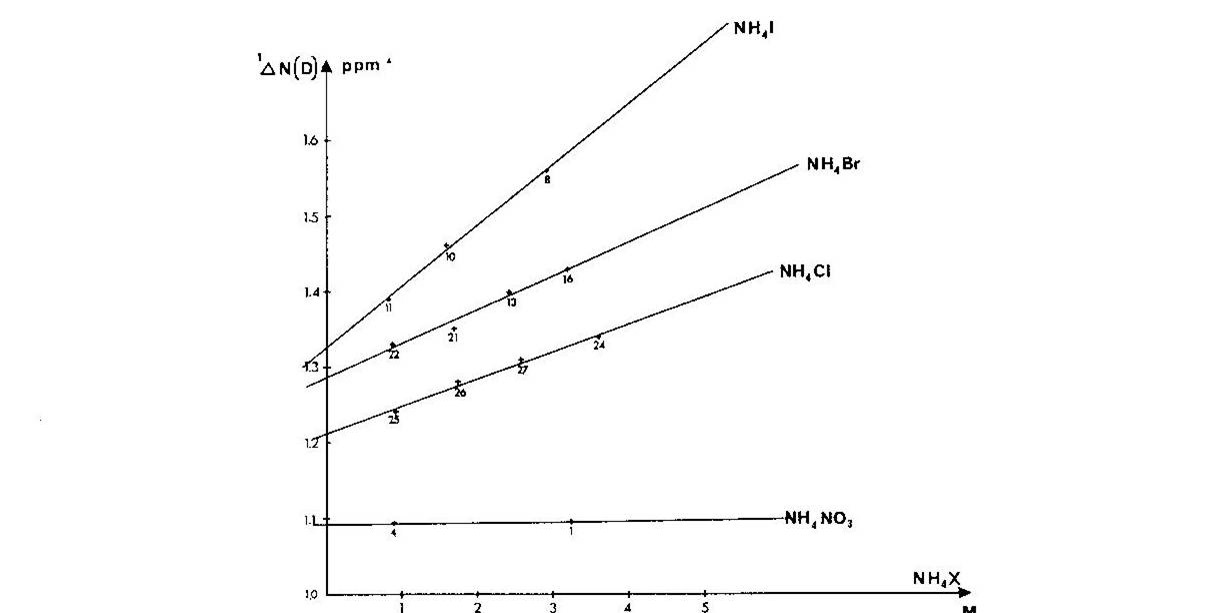
Ammonium ions are a close analogue of e.g.the side chain of protonated lysine. It was shown that one-bond deuterium isotope effects on 15N chemical shifts, 1ΔN(D), depended strongly on the counter ion, the counter ion concentration and the smallest 1ΔN(D) were found in very dilute solutions Fig. 4. A similar picture was seen for 2ΔH(D). [8]
Figure 4. Plot of one-bond deuterium isotope effects on 15N chemical shifts vs. concentration of ammonium salt. Taken from Ref. 7 with permission from the Danish Chemical Society
Furthermore, if the counter-ion was far away, ammonium ion buried in a cryptand like SC-24, no effect of the counter ion was found. [9] In addition, a solid state study showed that 1ΔN(D) decreases as the heavy atom distance decreases [10] and this was confirmed by theoretical calculations. [11] To sum up, 1ΔN(D) will be smaller, if a salt bridge is present, whereas 2ΔH(D) will become more negative.
5. Salt Bridges in Proteins
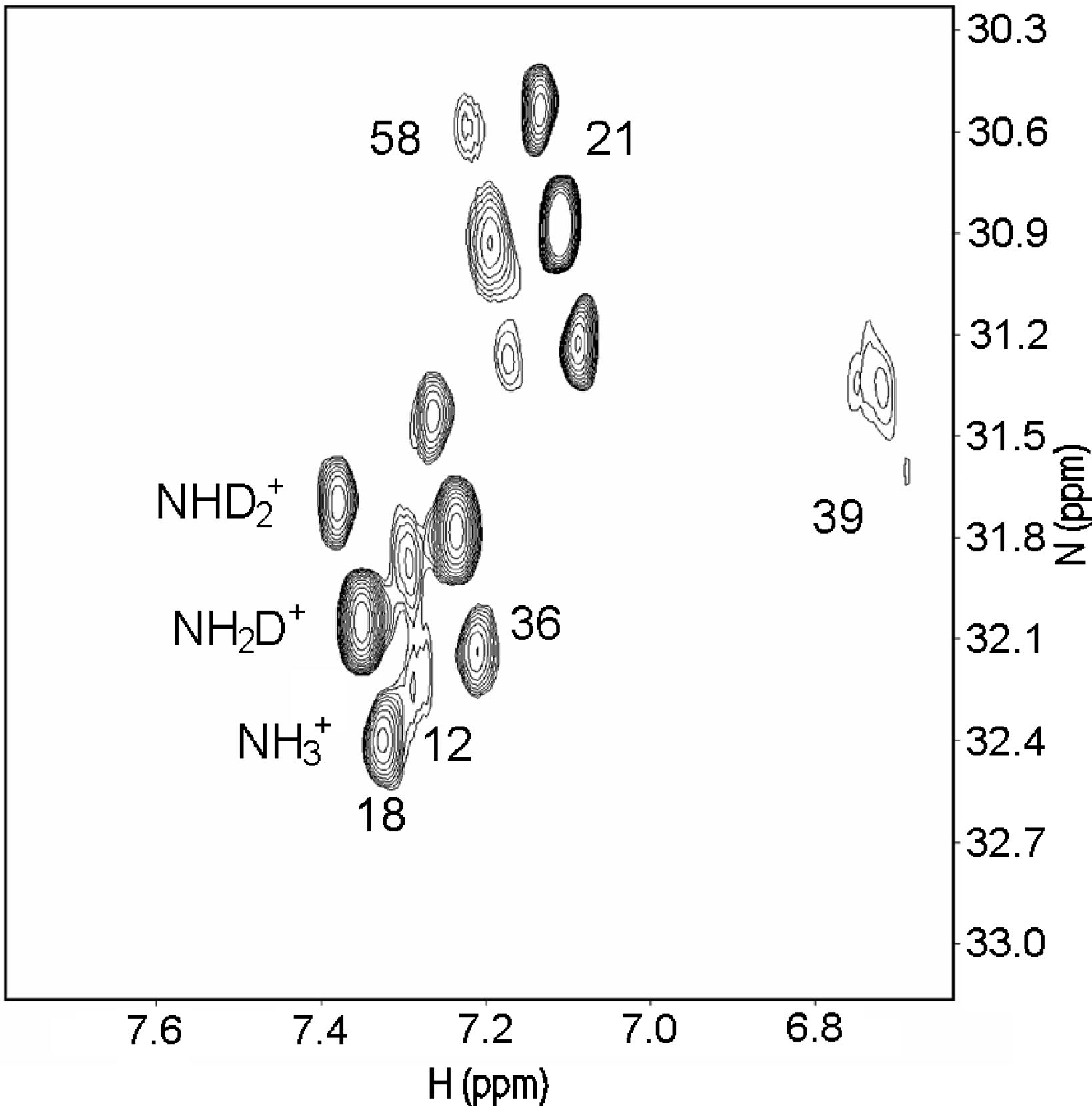
Salt bridges can both be "buried", in the inner part of the protein, or sovent exposed at the surface. Exchange of the NH protons can occur. A useful pulse sequence to overcome exchange problems is , HISQC [12]A typical spectrum is seen in Fig. 5.
Figure 5. HISQC spectrum of lysine NH3+ groups of protein G B1 domain at pH 5.45, 278 K in 40% D2O. Three peaks are seen for the lysines 18, 21, 36, and 58, corresponding to NH3+, NH2D+, and NHD2+groups (bottom right to top left, respectively: see labels on lysine18). From Ref. 5 with permission from the American Chemical Society.
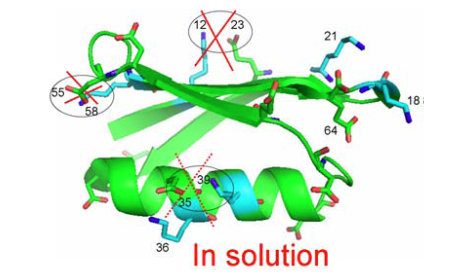
From Fig. 5 as well 1ΔN(D) as 2ΔH(D) isotope effects can be measured. The isotope effects turned out to be very similar for both what the crystal structure had indicated could be salt bridges and those not. So the conclusion as seen in Fig. 6 is that no salt bridges is to be found in protein G B1 domain.
Figure 6. Crystal structure of protein G B1 domain. The three salt bridges 35-36, 55-58 and 12-23 are crossed over as they do not exist in solution. From Ref. 5 with permission from the American Chemical Society
In contrast to protein G, salt bridges could be established in barnase in solution. Lysine 27 forms a salt bridge to Aspartic acid 54. Furthermore, if Arg 69 is mutated to Lys69, the latter also forms a salt bridge. Especially 2ΔH(D) is a good indicator as this becomes more negative when forming a salt bridge. In the absence of a salt bridge 1ΔN(D), 2ΔH(D):0.363 +-0.004 ppm, -0.026+-0.003 ppm, whereas in the presence of a salt bridge: 0.352+-0.004 ppm, -0.033+-0.002 ppm. [13]
Salt bridges may also involve arginines. Isotope effects were measured in the L99 mutant of T4 lysozyme. [14]
Iwahara et al. [12] demonstrated a salt bridge between lysine side-chains and the phosphate groups of DNA in the homeodomain-DNA complex also observing a 1ΔN(D).
5.1. Theoretical calculation of isotope effects on chemical shifts
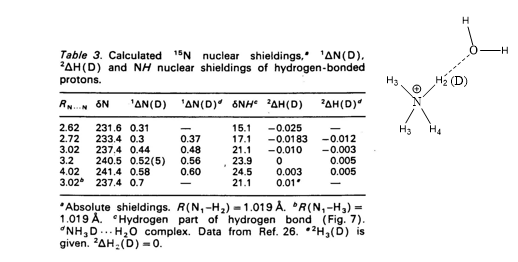
Theoretical calculations of isotope effects on chemical shifts can be based on the Jameson approach. [15] For ammonium ions this is shown in Table 1. The isotope effects on chemical shifts are calculated by shortening the NH bond length mimicking a ND bond.
Table 1. 1ΔN(D) and 2ΔH(D) of deuteriated ammonium ion complexed either with ammonia or water molecule (figure). Taken from Ref. 8 with permission from the Danish Chemical Society.
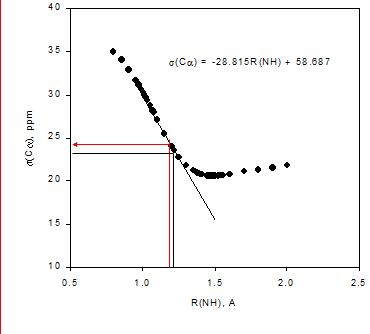
From Table 1 it is seen that 1ΔN(D) decreases as either ammonia or water is moving closer to the ammonium and for 2ΔH(D) even a change in sign is seen. This approach has also been used for protein G B1 domain. [5] A more recent approach is to calculate isotope effects by a direct treatment of the H/D effect using a multicomponent ab initio molecular orbital method. [16]
References
- K. D. Berndt, P. Guntert, L.P.M. Orbons, K. Wüthrich, Determination of a High-quality Nuclear Magnetic Resonance Solution Structure of the Bovine Pancreatic Trypsin Inhibitor and Comparison with Three Crystal Structures. J. Mol. Biol. 227 (1992) 757-775
- H. R. Bosshard, D. N. Marti and I. Jelesarov. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings J. Mol. Recognit. 17 (2004) 1–16. DOI:10.1002/jmr.657
- C. Lauritzen, O. Skovgaard, P. E. Hansen, and E. Tüchsen, Effects of N-terminal Extension Peptides on the Stucture and Stability of Bovine Pancreatic Trypsin Inhibitor Studied by 1H NMR. Int. J. Biol. Macromol., 14 (1992) 326-332.
- P. E. Hansen, W. Zhang, C. Lauritzen, S. Bjørn, L. C. Petersen, K. Norris, O. H. Olsen and Chr. Betzel, 13C NMR, X-ray and Differential Scanning Calorimetry Investigations of Truncated BPTI Mutants. Salt Bridge formation. Biochemistry 37 (1998) 3545-53.
- J.H. Tomlinson, S. Ullah, P. E. Hansen, M. P. Williamson. Characterisation of salt bridges to lysines in the Protein G B1 domain. J.Am.Chem.Soc. 131 (2009) 4674-84.
- P.W.D.Morgan,B.Birdsall,P.M.Nieto, A.R.Gargaro and J.Feeney, 1H/15N HSQC NMR Studies of Ligand Carboxylate Group Interactions with Arginine Residues in Complexes of Brodimoprim Analogues and Lactobacillus casei Dihydrofolate reductase. Biochemistry 38 (1999) 2127-2134
- P. E. Hansen, Isotope Effects on Chemical Shifts of Proteins and Peptides. Review. Magn.Reson.Chem. 38 (2000) 1-10.
- P. E. Hansen and A. Lycka, A Reinvestigation of One bond Deuterium Isotope Effects on Nitrogen and on Proton Nuclear Shielding for the Ammonium Ion. Acta Chem. Scand., 43 (1989) 222-232.
- P. E. Hansen, Aa. E. Hansen, A. Lycka and A. Buvari-Barcza, 2DeltaH(D) and 1DeltaN(D) Isotope Effects on Nuclear Shielding of Ammonium Ions in Complexes with Crown ethers and Cryptands. Acta Chem.Scand., 47 (1993) 777-788.
- P. E. Hansen, Deuterium Isotope Effects on 14,15N Chemical Shifts of Ammonium Ions. A Solid State NMR Study. Int.J.Inorg. Chem. (2011) https://doi.org/10.1155/2011/696497
- M. Munch, Aa. E. Hansen, P. E. Hansen and T. D. Bouman, Ab-Initio Calculations of Deuterium isotope Effects on hydrogen and Nitrogen nuclear Magnetic Shielding in the hydrated Ammonium Ion. Acta Chem. Scand., 46 (1992) 1065-1071.
- J. Iwahara, Y.-.S Jung, G. M. Clore. Heteronuclear NMR Spectroscopy for Lysine NH3Groupsin Proteins: Unique Effect of Water Exchange on 15N Transverse Relaxation. J.Am.Chem.Soc. 129 (2007) 2971-2980.
- M. P. Williamson, A. M. Hounslow, J. Ford, K. Fowler, M. Hebditch, P. E. Hansen, Detection of salt bridges to lysines in barnase in solution. Chem. Commun. 49 (2013) 9824-26.
- H.W.Mackenzie and D.F.Hansen, A 13C detected 15N double-quantum NMR experiment to probe arginine side-chain guanidinium 15Neta chemical shifts, J.Biomol. NMR 69 (2017) 123-132.
- C.J.Jameson, Isotopes in the Physical and Biomedical Sciences. Isotopic Applications in NMR Studies. E.Buncel and J.r.Jones, eds. Elsevier Science: Amsterdam, the Netherlands 1991; Vol.2.
- S. Ullah, T. Ishimoto, M.P.Williamson, P.E.Hansen, Ab Initio calculations of Deuterium isotope Effects on Chemical Shifts of Salt-bridged Lysines. J.Phys. Chem. B 115 (2011) 3208-3215.