You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Dean Liu and Version 1 by Ramendra Pati Pandey.
Biofilm has garnered a lot of interest due to concerns in various sectors such as public health, medicine, and the pharmaceutical industry. Biofilm-producing bacteria show a remarkable drug resistance capability, leading to an increase in morbidity and mortality.
- biofilm
- AMR
- silver nanoparticles
- multidrug resistance
- extracellular polymeric substances
1. Introduction
The existence of microbial pathogens has been recognized for many years, and since then, researchers have continuously tried to eliminate already existing and emerging pathogens causing infectious diseases, and develop antimicrobial agents to treat and eliminate the infectious disease. Antimicrobials represent a broad spectrum of compounds that may act against a wide repertoire of disease-causing microorganisms, such as bacteria, viruses, parasites, fungi, and protozoa [1]. These compounds have been used since the early 20th century to treat infected patients and have helped significantly in lowering the morbidity as well as mortality rates of most infectious diseases. In 1928, Alexander Fleming discovered penicillin, and in the 1940s, it came into clinical use just in time for World War II [2]. After only four years of use, the first penicillin-resistant strains of bacteria emerged; this resulted in the evolution of antimicrobial resistance (AMR). Subsequently, AMR has accelerated rapidly and expanded to different pathogenic species due to the persistent exposure and non-targeted application of antimicrobials in clinical and agricultural settings. Many current antimicrobials have become ineffective due to the advancement of AMR; it is believed that the non-judicious use and overdosing of antimicrobials are one of the foremost factors for the rise in drug-resistant bacteria [3]. Over last few decades, researchers have been trying to understand the mechanism of AMR and to develop new antimicrobials.
The ability to form biofilms is a universal attribute of bacteria. Biofilms are multicellular communities held together by a self-produced extracellular matrix. The mechanisms that a number of bacteria employ to form biofilms vary, frequently depending on environmental conditions and specific strain attributes. In the 17th century, Antonie van Leeuwenhoek first observed “animalcules” on his own teeth. In 1940, the “bottle effect” was observed in marine microorganisms [4]. This showed that the bacteria grow more often on the surface. Then, in 1943, biofilms were made by Zobell, and he found that bacteria on surfaces were greater in number compared with the surrounding seawater [5].
2. Mechanism of Biofilm Resistance
Antimicrobials (antibiotics, antivirals, antifungals, and antiparasitics) are compounds that kill microbes, stop their growth, and prevent or treat the infections in humans, animals, and plants. Antimicrobial resistance is the ability of microorganisms to survive against an antimicrobial drug at a higher concentration for a longer period, and is measured as minimum inhibitory concentration (MIC) [23][6]. Biofilm resistance can be antibiotic resistance or antibiotic tolerance. Microorganisms develop mechanisms against antimicrobials either through acquisition of foreign genetic material coding for resistant determinants by horizontal gene transfer (HGT) in biofilm EPS, or through genetic mutation (Figure 3). Mechanisms of AMR are: prevention of access or reduced permeability of antimicrobials, mutational changes in antibiotic targets, modification of targets, and enzymatic degradation of the antimicrobials by hydrolysis or chemical change (Figure 41). Antibiotic resistance (ABR) is a subdivision of AMR, as antibiotics are effective against the bacteria, but the bacteria become resistant to antibiotics.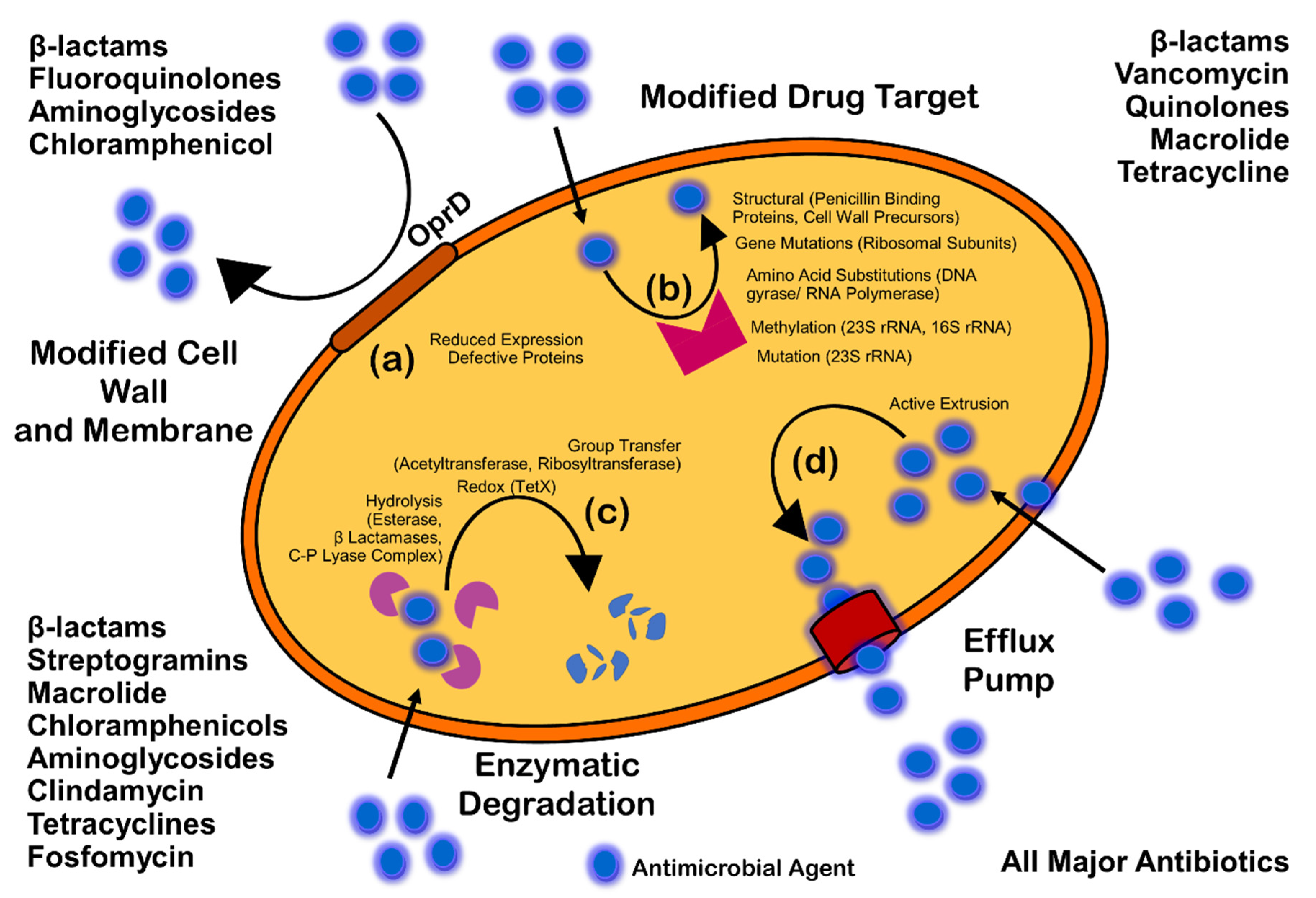
Figure 41. Mechanism of antimicrobial resistance: cell wall modification (a); modification of drug target (b); enzymatic degradation or destruction of antimicrobials (c); and efflux pump (d).
2.1. Prevention of Access or Reduced Penetration
The architecture, as well as composition, of ECM through gradients of dispersion can severely affect the penetration of antibiotics, the access to cells, and, finally, affect the efficacy of antibiotics. Diffusion of antibiotics also varies due to interaction with the ECM components [29][12]. For example, extracellular DNA enhances the resistance of Pseudomonas biofilm against aminoglycosides, but not against beta-lactams and fluoroquinolones [30,31,32][13][14][15]. In the same way, eDNA enhances the resistance of Staphylococcus epidermidis biofilm against glycopeptides. It has been seen that negatively charged eDNA binds to negatively charged glycopeptides (vancomycin) and aminoglycosides (tobramycin). It has also been demonstrated that the binding of vancomycin and eDNA is 100-fold higher than the vancomycin and D-Ala-D-Ala peptides in peptidoglycan precursors; this may result in the accumulation of eDNA in the ECM [32][15]. The retention of eDNA in the ECM may result in a cation-limited environment through the chelation of Mg2+ cations. The chelation of Mg2+ can also initiate the AMR-linked two-component system of PhoPQ and PmrAB for Psuedomonas aeruginosa and S. enterica serovar Typhimurium [31,33][14][16]. Additionally, the antibiotic modifying enzymes can be released and located in the ECM; these can also be used by other sensitive species of bacteria within a mixed species biofilm. For instance, beta lactamases released by Moraxella catarrhalis protect the S. pneumoniae and H. influenza against amoxicillin and ampicillin, respectively [34,35][17][18]. Therefore, the biofilm architecture can alter the diffusion of antibiotics, and also the exposure of cells.2.2. Stress Responses and Nutritional Limitation
Physiological heterogeneity is characterized by the genetic and phenotypic expression, metabolic activity, and antibiotic tolerance between the cells within different areas of biofilm [36,37][19][20]. The organization and architecture of biofilm generates gradients (Figure 3) of dispersion of water, nutrients, pH, signaling molecules, and waste products. It is believed that cells near the surface of biofilm microcolony utilize most of the nutrient supplies and create a deprived area further down [38,39,40,41][21][22][23][24]. Development of oxygen and nutritional depletion (Figure 3) in microcolonies and lower niches can contribute to the development of diverse physiological states of aerobic, anaerobic, microaerobic, and fermentative conditions; also the development of persisters, slow growth, fast growth, and dormant cells [17,36][19][25]. One such unique feature was observed by Yogesh and Anjali in 2021 (unpublished data), when MDR Enterococcus faecalis strains were re-cultured directly from the biofilm stage, which were stored for an extended period of 16 to 18 months at −70 °C. They found that the re-cultured colonies could grow only after 60–72 h of incubation at 37 °C on nutrient or chromogenic UTI agar without any supplement. When examined by the investigators of this study, the re-cultured bacterial strains were found to be resistant for the additional antibiotics. Cells in the oxygen deprived area (Figure 3) can show reduced metabolic activities and may undergo dormancy; this is believed to be the reason for tolerance against antibiotics such as tobramycin and ciprofloxacin that target protein synthesis and DNA gyrase [42][26]. It is well established that slow glowing cells are susceptible to colistin, which acts on the cell membrane [43][27]. However, the presence of colistin-tolerant cells within oxygen rich areas was observed, suggesting disagreement with the connection of slow growth rate and development of antibiotic tolerance within biofilm [44,45][28][29]. This fact was examined by Yogesh and Anjali in 2021 (unpublished data) when they found biofilm-producing Enterococcus faecalis resistant to colistin. Despite the full thickness antibiotic penetration, visible cellular activities and protein synthesis have also been seen in oxygen rich areas [46,47][30][31]. Through denitrification and fermentation, P. aeruginosa can sustain the anaerobic conditions, and supplementation of nitrate or L-arginine can increase the metabolic activity within nutrient deprived areas, thereby increasing the susceptibility to ciprofloxacin and tobramycin [42][26]. Stringent response (SR) and SOS response are adaptive mechanisms in response to stress and starvation of amino acids, iron, and carbon [48][32]. The tolerance of E. coli for cell wall inhibitor antibiotics such as carbapenems, penicillin, and cephalosporins is believed to be due to SR; this is also thought to be the case for cell division inhibitors such as norfloxacin and ofloxacin [49,50,51,52][33][34][35][36]. DNA damage may induce the SOS response and can initiate antibiotic tolerance. DNA damage leads to the generation of single-stranded DNA; that may activate RecA, stimulate self-cleavage of the repressor LexA and result in the de-repression of SOS genes [53][37]. The SOS response may lead to tolerance to antibiotics such as fluoroquinolones that can cause damage to DNA [54][38]. The tolerance of E. coli biofilm to fluoroquinolones has been observed due to the SOS response [52][36].2.3. Enzymatic Cell Wall Modification
The dlt genes are crucial for Enterococcus faecalis and Staphylococcus aureus to form biofilm [55,56][39][40]. This has been observed with the deletion of S. aureus dltA and the subsequent reduction of resistance to vancomycin, and also the reduction of planktonic resistance of E. faecalis to colistin and polymyxin B [57][41]. The dltABCD operon was a positive hit in a screening for biofilm-specific gentamicin tolerance genes in Streptococcus mutans, a dental pathogen that can also cause infective endocarditis [58][42]. The dltABCD homologues are important for D-alanylation of teichoic acid in many Gram-positive species [59][43]. Due to its inability to add D-alanine to the anionic teichoic acid molecules in the cell wall, the ΔdltA mutant was more negatively charged than the wild-type [58][42]. It is thought that the increased negative charge of the ΔdltA strain promotes uptake of gentamicin, a positively charged aminoglycoside [58][42].2.4. Multispecies Interaction
The interaction between different species (Figure 3) in biofilm can initiate antibiotic tolerance. For example, polymicrobial biofilms of E. faecalis, Finogoldia magna, and S. aureus were observed to be two-fold more resistant than the mono-species biofilm of P. aeruginosa [60][44]. In the same way, in a dual species biofilm model, M. catarhhalis released beta-lactamase, which protected S. pneumoniae from amoxicillin [34,61][17][45]. In the research by Ryan et al. (2008) on P. aeruginosa and Stenotrophomonas maltophilia dual-species biofilms, it was observed that the diffusible signal factor is an intercellular signaling molecule produced by S. maltophilia, and is sensed by the two-component sensor BptS in P. aeruginosa, leading to upregulation of the PmrA-regulated PA3552-3559 and PA4773-4775 genes [62][46]. Furthermore, the interaction between fungi and bacteria in a multispecies biofilm has also been studied. The resistance of Staphylococcus to vancomycin was increased in C. albicans and S. aureus biofilm due to the fungal matrix component beta-1,3-glucan, which is believed to act as a barrier against vancomycin [63,64][47][48]. It was also noted that C. albicans can increase the biofilm production of P. aeruginosa through alcohol production [65][49].2.5. Mutation
Genomic mutation, even without any strong spontaneous selective stress or pressure, may lead to AMR. Mutation usually occurs at the rate of 10−10 to 10−9 per nucleotide per generation in most of the bacteria [66,67][50][51]. Oxidative stress-causing agents that damage DNA can also accelerate the mutation rate. Although with a sublethal dose of a bactericidal, the build-up of ROS might be low but would be enough to induce synthesis of multidrug efflux pumps, mutagenesis, and promote resistance [68][52]. The defect in mutS, mutL, and uvrD genes can further promote the mutation frequency up to 100-fold due to failure of the DNA repair mechanism [69,70][53][54]. An example of evolutionary mutation in microorganisms can be seen with hypermutators with the capability to acquire favorable mutations under selective stress that can lead to AMR [71][55]. This particular phenotype has also been observed with Pseudomonas biofilm, with resistance against rifampicin and ciprofloxacin [72][56]. Apart from that the abovementioned phenotypic characteristics, hypermutations have also been reported in S. aureus and H. influenza isolated from cystic fibrosis infection but not in Enterobacteriaceae isolated from acute urinary tract infection; which indicates hypermutability is favored in certain environments [73,74,75][57][58][59]. Mutation in the rspL gene, the gene that codes 16S rDNA and S12 ribosomal protein, affects the antibiotic targeting for aminoglycosides, whereas mutation in the mexZ gene results in overproduction of the MexXY-OprM efflux system [76,77][60][61]. Additionally, the mutations in the genes coding for the PmrAB two-component regulatory system, which regulates the addition of aminoarabinose to lipid A, has been associated with colistin resistance [78][62]. Conversely, mutations in the promoter of the chromosomal ampC gene that increase the plasmid copy number may result in increased production of β-lactamases [79,80][63][64]. A large number of β-lactamase variants with point mutations in the gene, resulting in changes in the amino-acid sequence, may lead to the development of extended-spectrum β-lactamases (ESBLs) that also degrade first-, second-, and third-generation cephalosporins and/or became resistant to β-lactamase inhibitors [81][65].2.6. Efflux Pump
This is the movement of a drug from the intracellular to extracellular matrix without attachment to an intracellular target (Figure 3 and Figure 4); therefore, this mechanism confers resistance to bacterial cells [82][66]. Planktonic resistance in P. aeruginosa to low concentration ofloxacin has been said to be due to the multiple multidrug efflux pumps, such asMaxAB-OprM [83][67]. Another major multidrug efflux pump PA1875-1877 is believed to be a contributor to resistance in P. aeruginosa biofilm [84][68]. A two-fold to four-fold increase in susceptibility of biofilm to tobramycin, gentamicin, and ciprofloxacin was observed after the deletion of PA1875, PA1876, and PA1876; on the other hand, planktonic susceptibility was not affected much [84][68]. In addition, the resistance of P. aeruginosa biofilm to azithromycin was said to be due to the MexAB-OprM or MexCD-OprJ efflux pumps, whereas these efflux pumps are said to be required for the resistance against colistin in a metabolically active state [44,85][28][69].2.7. Quorum Sensing (QS)
QS is a population-density-dependent regulatory mechanism for interbacterial communication, which acts through signaling molecules named autoinducers. These autoinducers are recognized by the cell-surface receptors in order to induce gene transcription for virulence factors, surface proteins, transcriptional factors, and biofilm production [86,87][70][71]. A biofilm formed by P. aeruginosa lacking lasR and rhlR was observed as more susceptible to tobramycin than the wild-type biofilms [88][72]. In the same way, S. aureus deficient with QS-specific agrD was observed as less resistant as compared to the wild-type [89][73]. Moreover, fsrA and gelE mutants of E. faecalis for QS and QS-controlled protease were seen with the impairment of biofilm formation in the presence of daptomycin, gentamicin, or linezolid [90][74].References
- Burnett-Boothroyd, S.C.; McCarthy, B.J. 13-Antimicrobial treatments of textiles for hygiene and infection control applications: An Industrial perspective. In Textiles for Hygiene and Infection Control; McCarthy, B.J., Ed.; Woodhead Publishing Series in Textiles; Woodhead Publishing: Sawston, UK, 2011; pp. 196–209. ISBN 978-1-84569-636-8.
- Saga, T.; Yamaguchi, K. History of antimicrobial agents and resistant. Jpn. Med. Assoc. J. 2009, 52, 103–108.
- Ellis, A. Overcoming Resistance: A Rational Emotive Behavior Therapy Integrated Approach, 2nd ed.; Springer Publishing Company: Cham, Switzerland, 2002; ISBN 978-0-8261-4913-8.
- Percival, S.L.; Malic, S.; Cruz, H.; Williams, D.W. Introduction to biofilms. In Biofilms and Veterinary Medicine; Percival, S., Knottenbelt, D., Cochrane, C., Eds.; Springer Series on Biofilms; Springer: Berlin/Heidelberg, Germany, 2011; pp. 41–68. ISBN 978-3-642-21289-5.
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56.
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330.
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51.
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301.
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in gram-negative bacteria. Essays Biochem. 2017, 61, 49–59.
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448.
- Wilmaerts, D.; Windels, E.M.; Verstraeten, N.; Michiels, J. General mechanisms leading to persister formation and awakening. Trends Genet. TIG 2019, 35, 401–411.
- De Beer, D.; Stoodley, P.; Lewandowski, Z. Liquid flow in heterogeneous biofilms. Biotechnol. Bioeng. 1994, 44, 636–641.
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLOS Pathog. 2008, 4, e1000213.
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 60, 544–553.
- Doroshenko, N.; Tseng, B.S.; Howlin, R.P.; Deacon, J.; Wharton, J.A.; Thurner, P.J.; Gilmore, B.F.; Parsek, M.R.; Stoodley, P. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob. Agents Chemother. 2014, 58, 7273–7282.
- Johnson, L.; Horsman, S.R.; Charron-Mazenod, L.; Turnbull, A.L.; Mulcahy, H.; Surette, M.G.; Lewenza, S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella Enterica serovar typhimurium. BMC Microbiol. 2013, 13, 115.
- Perez, A.C.; Pang, B.; King, L.B.; Tan, L.; Murrah, K.A.; Reimche, J.L.; Wren, J.T.; Richardson, S.H.; Ghandi, U.; Swords, W.E. Residence of Streptococcus Pneumoniae and Moraxella Catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog. Dis. 2014, 70, 280–288.
- Armbruster, C.E.; Hong, W.; Pang, B.; Weimer, K.E.D.; Juneau, R.A.; Turner, J.; Swords, W.E. Indirect pathogenicity of haemophilus influenzae and Moraxella Catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 2010, 1, e00102-10.
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210.
- Williamson, K.S.; Richards, L.A.; Perez-Osorio, A.C.; Pitts, B.; McInnerney, K.; Stewart, P.S.; Franklin, M.J. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 2012, 194, 2062–2073.
- De Beer, D.; Stoodley, P.; Roe, F.; Lewandowski, Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 1994, 43, 1131–1138.
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella Pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824.
- Borriello, G.; Werner, E.; Roe, F.; Kim, A.M.; Ehrlich, G.D.; Stewart, P.S. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2004, 48, 2659–2664.
- Stewart, P.S.; Zhang, T.; Xu, R.; Pitts, B.; Walters, M.C.; Roe, F.; Kikhney, J.; Moter, A. Reaction–diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. npj Biofilms Microbiom. 2016, 2, 1–8.
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575.
- Borriello, G.; Richards, L.; Ehrlich, G.D.; Stewart, P.S. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2006, 50, 382–384.
- Haagensen, J.A.J.; Klausen, M.; Ernst, R.K.; Miller, S.I.; Folkesson, A.; Tolker-Nielsen, T.; Molin, S. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2007, 189, 28–37.
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the Pmr and MexAB-OprM genes. Mol. Microbiol. 2008, 68, 223–240.
- Chiang, W.-C.; Pamp, S.J.; Nilsson, M.; Givskov, M.; Tolker-Nielsen, T. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol. Med. Microbiol. 2012, 65, 245–256.
- Werner, E.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Heydorn, A.; Molin, S.; Pitts, B.; Stewart, P.S. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2004, 70, 6188–6196.
- Walters, M.C., III; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323.
- Kushwaha, G.S.; Oyeyemi, B.F.; Bhavesh, N.S. Stringent response protein as a potential target to intervene persistent bacterial infection. Biochimie 2019, 165, 67–75.
- Kusser, W.; Ishiguro, E.E. Suppression of mutations conferring penicillin tolerance by interference with the stringent control mechanism of Escherichia coli. J. Bacteriol. 1987, 169, 4396–4398.
- Tuomanen, E.; Tomasz, A. Induction of autolysis in nongrowing Escherichia coli. J. Bacteriol. 1986, 167, 1077–1080.
- Wu, N.; He, L.; Cui, P.; Wang, W.; Yuan, Y.; Liu, S.; Xu, T.; Zhang, S.; Wu, J.; Zhang, W.; et al. Ranking of persister genes in the same Escherichia coli genetic background demonstrates varying importance of individual persister genes in tolerance to different antibiotics. Front. Microbiol. 2015, 6, 3.
- Bernier, S.P.; Lebeaux, D.; DeFrancesco, A.S.; Valomon, A.; Soubigou, G.; Coppée, J.-Y.; Ghigo, J.-M.; Beloin, C. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013, 9, e1003144.
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384.
- Dörr, T.; Lewis, K.; Vulić, M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLOS Genet. 2009, 5, e1000760.
- Gross, M.; Cramton, S.E.; Götz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426.
- Fabretti, F.; Theilacker, C.; Baldassarri, L.; Kaczynski, Z.; Kropec, A.; Holst, O.; Huebner, J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 2006, 74, 4164–4171.
- Peschel, A.; Vuong, C.; Otto, M.; Götz, F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 2000, 44, 2845–2847.
- Nilsson, M.; Rybtke, M.; Givskov, M.; Høiby, N.; Twetman, S.; Tolker-Nielsen, T. The Dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int. J. Antimicrob. Agents 2016, 48, 298–304.
- Neuhaus, F.C.; Baddiley, J. A Continuum of anionic charge: Structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723.
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C.; Griswold, J.A.; Rumbaugh, K.P. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE 2011, 6, e27317.
- Budhani, R.K.; Struthers, J.K. Interaction of Streptococcus Pneumoniae and Moraxella Catarrhalis: Investigation of the indirect pathogenic role of β-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 1998, 42, 2521–2526.
- Ryan, R.P.; Fouhy, Y.; Garcia, B.F.; Watt, S.A.; Niehaus, K.; Yang, L.; Tolker-Nielsen, T.; Dow, J.M. Interspecies signalling via the Stenotrophomonas Maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 2008, 68, 75–86.
- Adam, B.; Baillie, G.S.; Douglas, L.J. Mixed species biofilms of Candida Albicans and Staphylococcus epidermidis. J. Med. Microbiol. 2002, 51, 344–349.
- Harriott, M.M.; Noverr, M.C. Candida Albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922.
- Chen, A.I.; Dolben, E.F.; Okegbe, C.; Harty, C.E.; Golub, Y.; Thao, S.; Ha, D.G.; Willger, S.D.; O’Toole, G.A.; Harwood, C.S.; et al. Candida Albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 2014, 10, e1004480.
- Woodford, N.; Ellington, M.J. The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 2007, 13, 5–18.
- Schroeder, J.W.; Yeesin, P.; Simmons, L.A.; Wang, J.D. Sources of spontaneous mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 29–48.
- Van Acker, H.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466.
- Schaaper, R.M.; Dunn, R.L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: The nature of in vivo DNA replication errors. Proc. Natl. Acad. Sci. USA 1987, 84, 6220–6224.
- Leong, P.M.; Hsia, H.C.; Miller, J.H. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J. Bacteriol. 1986, 168, 412–416.
- Blázquez, J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 37, 1201–1209.
- Driffield, K.; Miller, K.; Bostock, J.M.; O’Neill, A.J.; Chopra, I. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 2008, 61, 1053–1056.
- Prunier, A.-L.; Malbruny, B.; Laurans, M.; Brouard, J.; Duhamel, J.-F.; Leclercq, R. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 2003, 187, 1709–1716.
- Román, F.; Cantón, R.; Pérez-Vázquez, M.; Baquero, F.; Campos, J. Dynamics of long-term colonization of respiratory tract by haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 2004, 42, 1450–1459.
- Kovacs, B.; Le Gall-David, S.; Vincent, P.; Le Bars, H.; Buffet-Bataillon, S.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Is biofilm formation related to the hypermutator phenotype in clinical enterobacteriaceae isolates? FEMS Microbiol. Lett. 2013, 347, 116–122.
- Guénard, S.; Muller, C.; Monlezun, L.; Benas, P.; Broutin, I.; Jeannot, K.; Plésiat, P. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 58, 221–228.
- Springer, B.; Kidan, Y.G.; Prammananan, T.; Ellrott, K.; Böttger, E.C.; Sander, P. Mechanisms of streptomycin resistance: Selection of mutations in the 16S RRNA gene conferring resistance. Antimicrob. Agents Chemother. 2001, 45, 2877–2884.
- Moskowitz, S.M.; Ernst, R.K.; Miller, S.I. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 2004, 186, 575–579.
- Paltansing, S.; Kraakman, M.; van Boxtel, R.; Kors, I.; Wessels, E.; Goessens, W.; Tommassen, J.; Bernards, A. Increased expression levels of chromosomal AmpC β-lactamase in clinical Escherichia coli isolates and their effect on susceptibility to extended-spectrum cephalosporins. Microb. Drug Resist. 2015, 21, 7–16.
- van Boxtel, R.; Wattel, A.A.; Arenas, J.; Goessens, W.H.F.; Tommassen, J. Acquisition of carbapenem resistance by plasmid-encoded-AmpC-expressing Escherichia coli. Antimicrob. Agents Chemother. 2016, 61, e01413-16.
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended spectrum beta-lactamases: Definition, classification and epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–21.
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176.
- Brooun, A.; Liu, S.; Lewis, K. A Dose-Response Study of Antibiotic Resistance in Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2000, 44, 640–646.
- Zhang, L.; Mah, T.-F. Involvement of a Novel Efflux System in Biofilm-Specific Resistance to Antibiotics. J. Bacteriol. 2008, 190, 4447–4452.
- Gillis, R.J.; White, K.G.; Choi, K.-H.; Wagner, V.E.; Schweizer, H.P.; Iglewski, B.H. Molecular Basis of Azithromycin-Resistant Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2005, 49, 3858–3867.
- Di Cagno, R.; De Angelis, M.; Calasso, M.; Gobbetti, M. Proteomics of the bacterial cross-talk by quorum sensing. J. Proteom. 2011, 74, 19–34.
- Arevalo-Ferro, C.; Hentzer, M.; Reil, G.; Görg, A.; Kjelleberg, S.; Givskov, M.; Riedel, K.; Eberl, L. Identification of Quorum-Sensing Regulated Proteins in the Opportunistic Pathogen Pseudomonas aeruginosa by Proteomics. Environ. Microbiol. 2003, 5, 1350–1369.
- Bjarnsholt, T.; Jensen, P.Ø.; Burmølle, M.; Hentzer, M.; Haagensen, J.A.J.; Hougen, H.P.; Calum, H.; Madsen, K.G.; Moser, C.; Molin, S.; et al. Pseudomonas aeruginosa Tolerance to Tobramycin, Hydrogen Peroxide and Polymorphonuclear Leukocytes Is Quorum-Sensing Dependent. Microbiology 2005, 151, 373–383.
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850.
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.T.; Dunny, G.M. Multiple Roles for Enterococcus Faecalis Glycosyltransferases in Biofilm-Associated Antibiotic Resistance, Cell Envelope Integrity, and Conjugative Transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105.
More
