Celiac disease (CD) is an immune mediate disease characterised by gluten dependent T-cell mediated activation, autoimmunity and derangement of the intestinal mucosa in a specific genetic background. Although the activation of the T-cells has been studied in dept, the central question remains still unanswered, namely, why a pro-inflammatory T cell response to gluten is generated instead of a regulatory response, which normally promotes oral tolerance to dietary protein antigens.
Recent literature demonstrated tThat there is an inflamed environment in CD intestine, enriched in cytokines, such as IL-15, or type I interferons, in which T cells tend to acquire a pro- inflammatory phenotype. The factors that create a pro-inflammatory environment in the CD intestine, leading to an expansion of gliadin-specific T cells in genetically susceptible individuals and further shifting them towards a pro-inflammatory phenotype, remain to be identified. The recent literature is starting to address this question. From these studies it appears clearly, that subtle alterations on the CD cells are present also before the introduction of gluten. Gluten exacerbates these constitutive alterations, by increasing the same markers already altered before the gluten introduction, both in vitro and in vivo.
All these new observations add this disease “tout court” to the big family of increasing chronic inflammatory diseases where nutrients can have pro-inflammatory or anti-inflammatory effects, directly or indirectly mediated by the intestinal microbiota, where the intestine functions as a cross road for the control of the inflammation both local and at distance.
- celiac disease
- gluten
- inflammation
- microbiota
1. Celiac Disease as an Inflammatory Chronic Disease
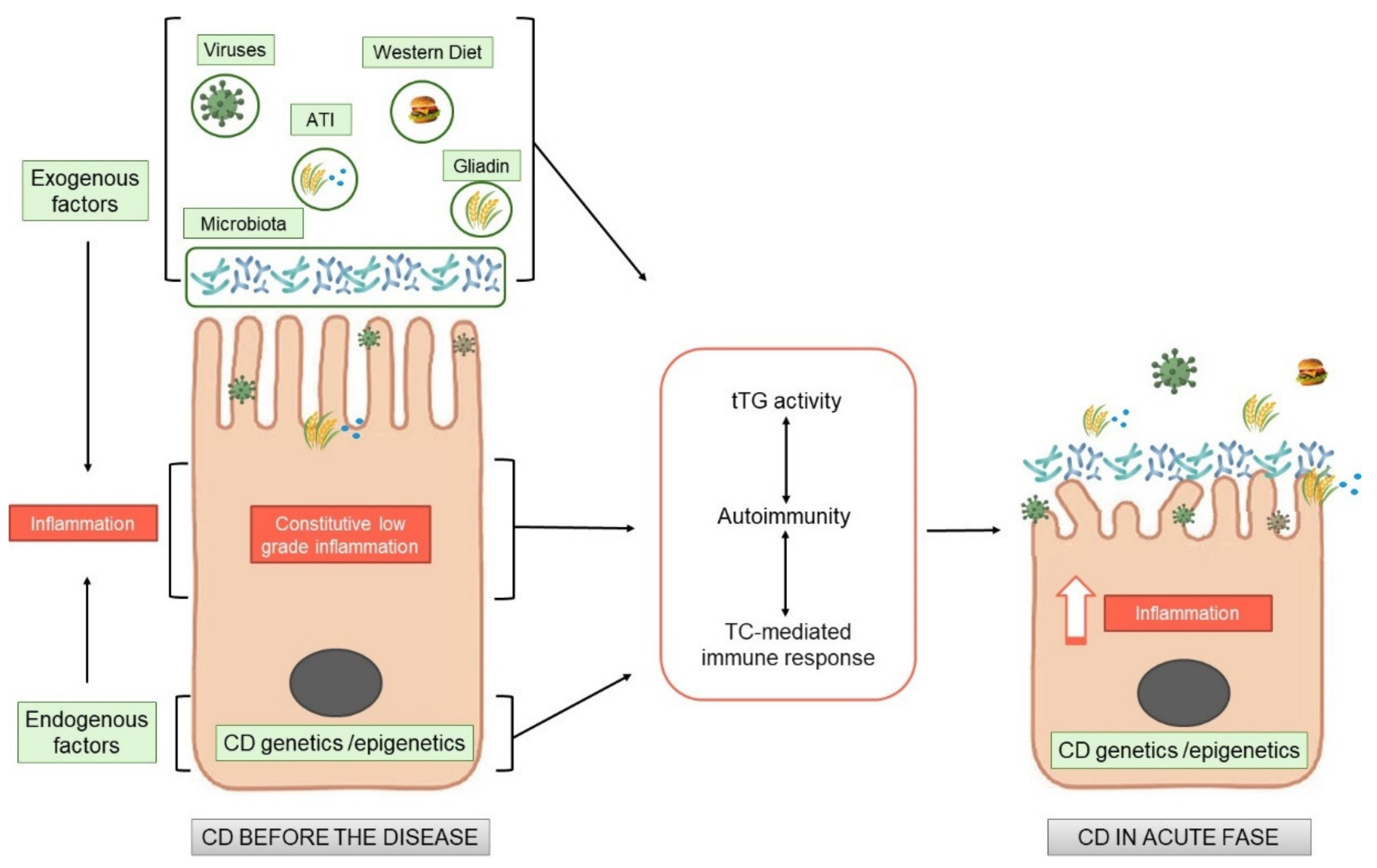
2. Endogenous Alterations in CD Independent of Gluten
| Endogenous Factors | |
|---|---|
| Models Investigated | Pathways Described |
| Children at risk of CD (before gluten introduction) |
|
| GFD–CD biopsies |
|
| Intestinal organoids derived from CD patients |
|
| Nonintestinal cells from GFD–CD patients |
|
2.1. Patients at Risk of CD: What Happens before the Disease?
2.2. GFD–CD Patient Biopsies, Lessons from In Vivo and In Vitro Studies
Celiac patients after diagnosis have to initiate a strict diet without gluten. These patients are defined as gluten-free diet–celiac disease patients (GFD–CD). The dietary restrictions have to be followed for life. GFD–CD patients do not eat gluten; thus, the main cause of the disease is not present, but some alterations mainly at the intestinal level can still be found.GFD–CD Patients before and after Gluten Challenge In Vivo
New observations have been obtained and published challenging gluten in vivo in GFD–CD patients by analyzing before and after gluten introduction on several different pathways such as proliferation, inflammation and differentiation at the intestinal level [25,26,27][9][10][11]. Dotsenko et al., demonstrated that the intestinal morphology in patients on a strict GFD was similar to that measured in control subjects [25][9]. However, gene transcription showed that the GFD–CD group differed significantly from the control group, showing a substantial number of differentially expressed genes. Gene Ontology and Reactome pathway analyses revealed that patients on a GFD presented altered expression of genes with functions such as brush border assembly, developmental processes, transport of small molecules, and FOXO (Forkhead box-O)-mediated transcription of cell cycle genes [25][9]. Moreover, the Wnt (Wingless and Int 1) pathway has been found altered in GFD–CD patients with respect to controls, suggesting that the alterations of a pathway nodal to intestinal differentiation and homeostasis is also present in the absence of gluten [25][9]. In the gastrointestinal tract, Wnt signaling activation drives homeostasis and damage-induced repair. Wnt signaling also has a key role during stem cell-driven intestinal homeostasis, regeneration, ageing and cancer [59][44]. After being gluten challenged, the Wnt pathway was further altered, as tested by RNA sec, in intestinal biopsies of patients on a GFD with respect to controls [25][9]. All together, these data show that even on a strict GFD, patients reveal patterns of ongoing disease and that gluten acts on the same pathways already altered. Another recent paper by Stamnaes J. et al. [26,27][10][11] shows by proteomics analysis that before being challenged in GFD–CD patients, epithelial inflammation is already in the intestines, with changes indicative of minor crypt hyperplasia and low-grade inflammation in the serum. After challenge with gluten, an increase in the same proteins, already altered at base line in GFD–CD responders, was found. Despite clinical and histological remission, celiac disease patients that develop a mucosal response after 14 days of gluten challenge have already at baseline altered protein compositions of their gut tissue with signs of ongoing inflammation. In conclusion, these observations indicate that in GFD–CD patients, several different pathways such as proliferation, inflammation and differentiation are altered at the intestinal level and that they can be further altered by gliadin challenge.GFD–CD Patient Biopsies
Several papers have been published studying the biopsies of GFD–CD patients. In these papers, several pathways have been found altered by different techniques. Most of the papers analyze the whole biopsies, as in the case of genetic or expression studies, while in some others, only the epithelium are evaluated. The following listed are the most significant literature contributions that we discuss, clustered by the pathways found altered.- A.
-
Inflammation
- B.
-
Innate immunity pathways
- C.
-
Enterocyte proliferation and differentiation
- D.
-
Structural alterations
2.3. Intestinal Organoids
An emerging role in CD pathogenesis has been attributed to the intestinal epithelium. In epithelial cells of CD patients, morphological and functional alterations have been described together with the activation of the inflammasome pathway [39,79][23][64]. Organoids derived from the small intestine represent a new tool to study the role of the intestinal epithelium in several different diseases [80][65]. Intestinal organoids are derived from crypt stem cells cultivated in 3D and embedded in a matrix; they resemble the small intestinal epithelium. Organoids from CD patients have shown the presence of increased staminality, permeability, inflammasome activity, and innate immunity genes with respect to organoids in healthy individuals [48,79][33][64]. Instead, extracellular matrix (ECM) genes were decreased in [49][34]. Moreover, increased markers of inflammation were found at the protein and mRNA levels in CD organoids. This inflammation was not a residual effect of the tissue of origin but is probably constitutive, as it was persistent even after many days in culture [17][1]. In CD biopsies and in intestinal organoids, increased sensitivity to inflammatory stimuli from bacteria [79][64], viral ligand loxoribine [17][1] and gliadin peptide P31-43 [17][1] have been described, indicating that intestinal organoids from CD patients are more sensitive to pro-inflammatory stimuli. Taken all together, these observations indicate that CD intestinal epithelial cells are constitutively different from those in healthy individuals.
References
- Porpora, M.; Conte, M.; Lania, G.; Bellomo, C.; Rapacciuolo, L.; Chirdo, F.G.; Auricchio, R.; Troncone, R.; Auricchio, S.; Barone, M.V.; et al. Inflammation Is Present, Persistent and More Sensitive to Proinflammatory Triggers in Celiac Disease Enterocytes. Int. J. Mol. Sci. 2022, 23, 1973.
- Auricchio, R.; Troncone, R. Can Celiac Disease Be Prevented? Front. Immunol. 2021, 12, 672148.
- Auricchio, R.; Galatola, M.; Cielo, D.; Amoresano, A.; Caterino, M.; De Vita, E.; Illiano, A.; Troncone, R.; Greco, L.; Ruoppolo, M. A Phospholipid Profile at 4 Months Predicts the Onset of Celiac Disease in at-Risk Infants. Sci. Rep. 2019, 9, 14303.
- Sen, P.; Carlsson, C.; Virtanen, S.M.; Simell, S.; Hyöty, H.; Ilonen, J.; Toppari, J.; Veijola, R.; Hyötyläinen, T.; Knip, M.; et al. Persistent Alterations in Plasma Lipid Profiles Before Introduction of Gluten in the Diet Associated with Progression to Celiac Disease. Clin. Transl. Gastroenterol. 2019, 10, 1–10.
- Auricchio, R.; Stellato, P.; Bruzzese, D.; Cielo, D.; Chiurazzi, A.; Galatola, M.; Castilljeo, G.; Crespo Escobar, P.; Gyimesi, J.; Hartman, C.; et al. Growth rate of coeliac children is compromised before the onset of the disease. Arch. Dis. Child. 2020, 105, 964–968.
- Auricchio, R.; Calabrese, I.; Galatola, M.; Cielo, D.; Carbone, F.; Mancuso, M.; Matarese, G.; Troncone, R.; Auricchio, S.; Greco, L. Author Correction: Gluten consumption and inflammation affect the development of celiac disease in at-risk children. Sci. Rep. 2022, 12, 8157.
- Galatola, M.; Cielo, D.; Panico, C.; Stellato, P.; Malamisura, B.; Carbone, L.; Gianfrani, C.; Troncone, R.; Greco, L.; Auricchio, R. Presymptomatic Diagnosis of Celiac Disease in Predisposed Children: The Role of Gene Expression Profile. J. Pediatr. Gastroenterol. Nutr. 2017, 65, 314–320.
- Olivares, M.; Walker, A.W.; Capilla, A.; Benítez-Páez, A.; Palau, F.; Parkhill, J.; Castillejo, G.; Sanz, Y. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome 2018, 6, 36.
- Dotsenko, V.; Oittinen, M.; Taavela, J.; Popp, A.; Peräaho, M.; Staff, S.; Sarin, J.; Leon, F.; Isola, J.; Mäki, M.; et al. Genome-Wide Transcriptomic Analysis of Intestinal Mucosa in Celiac Disease Patients on a Gluten-Free Diet and Postgluten Challenge. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 13–32.
- Stamnaes, J.; Stray, D.; Stensland, M.; Sarna, V.K.; Nyman, T.A.; Lundin, K.E.A.; Sollid, L.M. Quantitative proteomics of coeliac gut during 14-day gluten challenge: Low-level baseline inflammation despite clinical and histological normality predicts subsequent response. medRxiv 2020.
- Stamnaes, J.; Stray, D.; Stensland, M.; Sarna, V.K.; Nyman, T.A.; Lundin, K.E.A.; Sollid, L.M. In Well-Treated Celiac Patients Low-Level Mucosal Inflammation Predicts Response to 14-day Gluten Challenge. Adv. Sci. 2021, 8, 2003526.
- Trynka, G.; Zhernakova, A.; Romanos, J.; Franke, L.; Hunt, K.A.; Turner, G.; Bruinenberg, M.; Heap, G.A.; Platteel, M.; Ryan, A.W.; et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut 2009, 58, 1078–1083.
- Fernandez-Jimenez, N.; Castellanos-Rubio, A.; Plaza-Izurieta, L.; Irastorza, I.; Elcoroaristizabal, X.; Jauregi-Miguel, A.; Lopez-Euba, T.; Tutau, C.; de Pancorbo, M.M.; Vitoria, J.C.; et al. Coregulation and modulation of NFκB-related genes in celiac disease: Uncovered aspects of gut mucosal inflammation. Hum. Mol. Genet. 2014, 23, 1298–1310.
- Castellanos-Rubio, A.; Bilbao, J.R. Profiling Celiac Disease-Related Transcriptional Changes. Int. Rev. Cell. Mol. Biol. 2018, 336, 149–174.
- Castellanos-Rubio, A.; Ghosh, S. Disease-Associated SNPs in Inflammation-Related lncRNAs. Front. Immunol. 2019, 10, 420.
- Jabri, B.; Sollid, L.M. Mechanisms of disease: Immunopathogenesis of celiac disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 516–525.
- Sziksz, E.; Veres, G.; Vannay, A.; Prókai, A.; Gál, K.; Onody, A.; Korponay-Szabó, I.R.; Reusz, G.; Szabó, A.; Tulassay, T.; et al. Increased heat shock protein 72 expression in celiac disease. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 573–578.
- Abadie, V.; Jabri, B. IL-15: A central regulator of celiac disease immunopathology. Immunol. Rev. 2014, 260, 221–234.
- Bernardo, D.; Garrote, J.A.; Allegretti, Y.; León, A.; Gómez, E.; Bermejo-Martin, J.F.; Calvo, C.; Riestra, S.; Fernández-Salazar, L.; Blanco-Quirós, A.; et al. Higher constitutive IL15R alpha expression and lower IL-15 response threshold in coeliac disease patients. Clin. Exp. Immunol. 2008, 154, 64–73.
- Nanayakkara, M.; Lania, G.; Maglio, M.; Auricchio, R.; De Musis, C.; Discepolo, V.; Miele, E.; Jabri, B.; Troncone, R.; Auricchio, S.; et al. P31-43, an undigested gliadin peptide, mimics and enhances the innate immune response to viruses and interferes with endocytic trafficking: A role in celiac disease. Sci. Rep. 2018, 8, 10821.
- Allard-Chamard, H.; Mishra, H.K.; Nandi, M.; Mayhue, M.; Menendez, A.; Ilangumaran, S.; Ramanathan, S. Interleukin-15 in autoimmunity. Cytokine 2020, 136, 155258.
- Abadie, V.; Kim, S.M.; Lejeune, T.; Palanski, B.A.; Ernest, J.D.; Tastet, O.; Voisine, J.; Discepolo, V.; Marietta, E.V.; Hawash, M.B.F.; et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature 2020, 578, 600–604.
- Lania, G.; Nanayakkara, M.; Maglio, M.; Auricchio, R.; Porpora, M.; Conte, M.; De Matteis, M.A.; Rizzo, R.; Luini, A.; Discepolo, V.; et al. Constitutive alterations in vesicular trafficking increase the sensitivity of cells from celiac disease patients to gliadin. Commun. Biol. 2019, 2, 190.
- Barone, M.V.; Troncone, R.; Auricchio, S. Gliadin peptides as triggers of the proliferative and stress/innate immune response of the celiac small intestinal mucosa. Int. J. Mol. Sci. 2014, 15, 20518–20537.
- Juuti-Uusitalo, K.; Mäki, M.; Kainulainen, H.; Isola, J.; Kaukinen, K. Gluten affects epithelial differentiation-associated genes in small intestinal mucosa of coeliac patients. Clin. Exp. Immunol. 2007, 150, 294–305.
- Nanayakkara, M.; Lania, G.; Maglio, M.; Kosova, R.; Sarno, M.; Gaito, A.; Discepolo, V.; Troncone, R.; Auricchio, S.; Auricchio, R.; et al. Enterocyte proliferation and signaling are constitutively altered in celiac disease. PLoS ONE 2013, 8, e76006.
- Thorburn, A.N.; Macia, L.; Mackay, C.R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014, 40, 833–842.
- Bjarnason, I.; Marsh, M.N.; Price, A.; Levi, A.J.; Peters, T.J. Intestinal permeability in patients with coeliac disease and dermatitis herpetiformis. Gut 1985, 26, 1214–1219.
- Bjarnason, I.; Peters, T.J. In vitro determination of small intestinal permeability: Demonstration of a persistent defect in patients with coeliac disease. Gut 1984, 25, 145–150.
- Ciccocioppo, R.; Finamore, A.; Ara, C.; Di Sabatino, A.; Mengheri, E.; Corazza, G.R. Altered expression, localization, and phosphorylation of epithelial junctional proteins in celiac disease. Am. J. Clin. Pathol. 2006, 125, 502–511.
- Schulzke, J.D.; Bentzel, C.J.; Schulzke, I.; Riecken, E.O.; Fromm, M. Epithelial tight junction structure in the jejunum of children with acute and treated celiac sprue. Pediatr. Res. 1998, 43, 435–441.
- Jauregi-Miguel, A.; Fernandez-Jimenez, N.; Irastorza, I.; Plaza-Izurieta, L.; Vitoria, J.C.; Bilbao, J.R. Alteration of tight junction gene expression in celiac disease. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 762–767.
- Freire, R.; Ingano, L.; Serena, G.; Cetinbas, M.; Anselmo, A.; Sapone, A.; Sadreyev, R.I.; Fasano, A.; Senger, S. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 2019, 9, 7029.
- Dieterich, W.; Neurath, M.F.; Zopf, Y. Intestinal ex vivo organoid culture reveals altered programmed crypt stem cells in patients with celiac disease. Sci. Rep. 2020, 10, 3535.
- Nanayakkara, M.; Kosova, R.; Lania, G.; Sarno, M.; Gaito, A.; Galatola, M.; Greco, L.; Cuomo, M.; Troncone, R.; Auricchio, S.; et al. A celiac cellular phenotype, with altered LPP sub-cellular distribution, is inducible in controls by the toxic gliadin peptide P31-43. PLoS ONE 2013, 8, e79763.
- Paolella, G.; Nanayakkara, M.; Sposito, S.; Lepretti, M.; Auricchio, S.; Esposito, C.; Barone, M.V.; Martucciello, S.; Caputo, I. Constitutive Differential Features of Type 2 Transglutaminase in Cells Derived from Celiac Patients and from Healthy Subjects. Int. J. Mol. Sci. 2020, 21, 1231.
- Discepolo, V.; Lania, G.; Ten Eikelder, M.L.G.; Nanayakkara, M.; Sepe, L.; Tufano, R.; Troncone, R.; Auricchio, S.; Auricchio, R.; Paolella, G.; et al. Pediatric Celiac Disease Patients Show Alterations of Dendritic Cell Shape and Actin Rearrangement. Int. J. Mol. Sci. 2021, 22, 2708.
- Vriezinga, S.L.; Auricchio, R.; Bravi, E.; Castillejo, G.; Chmielewska, A.; Crespo Escobar, P.; Kolaček, S.; Koletzko, S.; Korponay-Szabo, I.R.; Mummert, E.; et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 2014, 371, 1304–1315.
- Chirdo, F.G.; Auricchio, S.; Troncone, R.; Barone, M.V. The gliadin P31-43 peptide: Inducer of multiple proinflammatory effects. Int. Rev. Cell Mol. Biol. 2021, 358, 165–205.
- Barone, M.V.; Nanayakkara, M.; Paolella, G.; Maglio, M.; Vitale, V.; Troiano, R.; Ribecco, M.T.; Lania, G.; Zanzi, D.; Santagata, S.; et al. Gliadin peptide P31-43 localises to endocytic vesicles and interferes with their maturation. PLoS ONE 2010, 5, e12246.
- Wenk, M.R. Lipidomics: New tools and applications. Cell 2010, 143, 888–895.
- Oresic, M.; Simell, S.; Sysi-Aho, M.; Näntö-Salonen, K.; Seppänen-Laakso, T.; Parikka, V.; Katajamaa, M.; Hekkala, A.; Mattila, I.; Keskinen, P.; et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J. Exp. Med. 2008, 205, 2975–2984.
- Lamichhane, S.; Ahonen, L.; Dyrlund, T.S.; Kemppainen, E.; Siljander, H.; Hyöty, H.; Ilonen, J.; Toppari, J.; Veijola, R.; Hyötyläinen, T.; et al. Dynamics of Plasma Lipidome in Progression to Islet Autoimmunity and Type 1 Diabetes—Type 1 Diabetes Prediction and Prevention Study (DIPP). Sci. Rep. 2018, 8, 10635.
- Perochon, J.; Carroll, L.R.; Cordero, J.B. Wnt Signalling in Intestinal Stem Cells: Lessons from Mice and Flies. Genes 2018, 9, 138.
- Hayden, M.S.; Ghosh, S. NF-κB in immunobiology. Cell. Res. 2011, 21, 223–244.
- Barone, M.V.; Auricchio, S. A Cumulative Effect of Food and Viruses to Trigger Celiac Disease (CD): A Commentary on the Recent Literature. Int. J. Mol. Sci. 2021, 22, 2027.
- Marsh, M.; Loft, D.; Garner, V.; Gordon, D. Time/dose responses of coeliac mucosae to graded oral challenges with Frazer’s fraction III of gliadin. Eur. J. Gastroenterol. Hepatol. 1992, 4, 667–673.
- Mulder, C.J.; Mearin, M.L.; Peña, A.S. Clinical and pathological spectrum of coeliac disease. Gut 1993, 34, 1740–1741.
- Marsh, M.N.; Crowe, P.T. Morphology of the mucosal lesion in gluten sensitivity. Baillieres Clin. Gastroenterol. 1995, 9, 273–293.
- Barone, M.V.; Gimigliano, A.; Castoria, G.; Paolella, G.; Maurano, F.; Paparo, F.; Maglio, M.; Mineo, A.; Miele, E.; Nanayakkara, M.; et al. Growth factor-like activity of gliadin, an alimentary protein: Implications for coeliac disease. Gut 2007, 56, 480–488.
- Taavela, J.; Viiri, K.; Popp, A.; Oittinen, M.; Dotsenko, V.; Peräaho, M.; Staff, S.; Sarin, J.; Leon, F.; Mäki, M.; et al. Histological, immunohistochemical and mRNA gene expression responses in coeliac disease patients challenged with gluten using PAXgene fixed paraffin-embedded duodenal biopsies. BMC Gastroenterol. 2019, 19, 189.
- Cervio, E.; Volta, U.; Verri, M.; Boschi, F.; Pastoris, O.; Granito, A.; Barbara, G.; Parisi, C.; Felicani, C.; Tonini, M.; et al. Sera of patients with celiac disease and neurologic disorders evoke a mitochondrial-dependent apoptosis in vitro. Gastroenterology 2007, 133, 195–206.
- Volta, U.; De Giorgio, R.; Granito, A.; Stanghellini, V.; Barbara, G.; Avoni, P.; Liguori, R.; Petrolini, N.; Fiorini, E.; Montagna, P.; et al. Anti-ganglioside antibodies in coeliac disease with neurological disorders. Dig. Liver. Dis. 2006, 38, 183–187.
- Barone, M.V.; Caputo, I.; Ribecco, M.T.; Maglio, M.; Marzari, R.; Sblattero, D.; Troncone, R.; Auricchio, S.; Esposito, C. Humoral immune response to tissue transglutaminase is related to epithelial cell proliferation in celiac disease. Gastroenterology 2007, 132, 1245–1253.
- Granito, A.; Muratori, P.; Cassani, F.; Pappas, G.; Muratori, L.; Agostinelli, D.; Veronesi, L.; Bortolotti, R.; Petrolini, N.; Bianchi, F.B.; et al. Anti-actin IgA antibodies in severe coeliac disease. Clin. Exp. Immunol. 2004, 137, 386–392.
- Zauli, D.; Grassi, A.; Granito, A.; Foderaro, S.; De Franceschi, L.; Ballardini, G.; Bianchi, F.B.; Volta, U. Prevalence of silent coeliac disease in atopics. Dig. Liver Dis. 2000, 32, 775–779.
- Wapenaar, M.C.; Monsuur, A.J.; van Bodegraven, A.A.; Weersma, R.K.; Bevova, M.R.; Linskens, R.K.; Howdle, P.; Holmes, G.; Mulder, C.J.; Dijkstra, G.; et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut 2008, 57, 463–467.
- Trynka, G.; Hunt, K.A.; Bockett, N.A.; Romanos, J.; Mistry, V.; Szperl, A.; Bakker, S.F.; Bardella, M.T.; Bhaw-Rosun, L.; Castillejo, G.; et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011, 43, 1193–1201.
- Kaminsky, L.W.; Al-Sadi, R.; Ma, T.Y. IL-1β and the Intestinal Epithelial Tight Junction Barrier. Front. Immunol. 2021, 12, 767456.
- De Matteis, M.A.; Luini, A. Mendelian disorders of membrane trafficking. N. Engl. J. Med. 2011, 365, 927–938.
- Adolph, T.E.; Tomczak, M.F.; Niederreiter, L.; Ko, H.J.; Böck, J.; Martinez-Naves, E.; Glickman, J.N.; Tschurtschenthaler, M.; Hartwig, J.; Hosomi, S.; et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013, 503, 272–276.
- Watkin, L.B.; Jessen, B.; Wiszniewski, W.; Vece, T.J.; Jan, M.; Sha, Y.; Thamsen, M.; Santos-Cortez, R.L.; Lee, K.; Gambin, T.; et al. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat. Genet. 2015, 47, 654–660.
- Fernandez-Jimenez, N.; Garcia-Etxebarria, K.; Plaza-Izurieta, L.; Romero-Garmendia, I.; Jauregi-Miguel, A.; Legarda, M.; Ecsedi, S.; Castellanos-Rubio, A.; Cahais, V.; Cuenin, C.; et al. The methylome of the celiac intestinal epithelium harbours genotype-independent alterations in the HLA region. Sci. Rep. 2019, 9, 1298.
- Pietz, G.; De, R.; Hedberg, M.; Sjöberg, V.; Sandström, O.; Hernell, O.; Hammarström, S.; Hammarström, M.L. Immunopathology of childhood celiac disease-Key role of intestinal epithelial cells. PLoS ONE 2017, 12, e0185025.
- Li, V.S.W. Modelling intestinal inflammation and infection using ‘mini-gut’ organoids. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 89–90.
