The coronavirus disease 19 (COVID-19) is caused by the highly transmissible severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has affected the global population despite socioeconomic status and amazed surveillance agencies for its incidence, mortality, and recovery rates. COVID-19 affects all age groups; however, it is suggested to progress into severe disease and cause mortality in over 10% of the confirmed cases, depending on the individual characteristics of the affected population. One of the biggest unanswered questions it is why only some individuals develop into the severe stages of the disease. Current data indicate that most of the critically ill are the elderly or those with comorbidities such as hypertension, diabetes, and asthma. However, it has been noted that, in some populations, severe disease is mostly observed in much younger individuals (<60-years old) with no reported underlying medical conditions. Certainly, many factors may contribute to disease severity including intrinsic host factors such as genetic variants, the expression levels of tissue proteins, among others.
- COVID-19
- SARS-CoV-2
- host factors
- tissue proteases
- immune responses
- disease outcome
Primarily defined as a disease of the respiratory tract, SARS-CoV-2 infection is now known to cause a systemic pathogenesis, targeting multiple organs and ultimately leading to their failure and to patient death (Figure 1). Disease transmission is still uncertain, but it is thought to happen through direct contact, respiratory droplets, or aerosols, and even by ingestion of viral particles [21][1]. Once transmitted to an individual, the virus enters the human cells via a direct interaction of its spike protein with the host ACE2. ACE2 is highly expressed not only in human alveolar epithelial cells, but also in various others including the tongue and oral mucosa epithelial cells, leukocytes, blood vessels, heart, kidney, endothelium, and intestine [22–24][2][3][4]. Other broadly expressed host proteins contribute to an effective infection of SARS-CoV-2 into human cells including the proteases furin and furin-like proteins [25][5], transmembrane protease serine 2 (TMPRSS2), and the cathepsin B and L (CatB/L) [26][6]. At this early phase of infection, the expression profiles of these proteins as well as interferon (IFN) production are determinant pathways for disease outcome. As disease progresses, cellular immune responses become essential players in the host fight to SARS-CoV-2. Imbalances in these pathways such as defective IFN production or ineffective cell migration/activation may, therefore, result in a worsened disease prognosis.
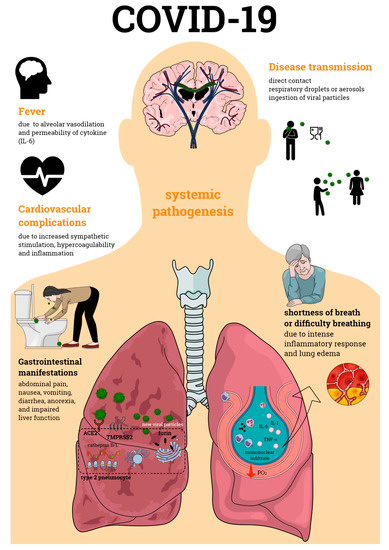
Figure 1. Coronavirus disease 19 (COVID-19) pathogenesis. COVID-19 is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which primarily affects the respiratory tract causing bilateral pneumonia. Its transmission may occur through direct contact and respiratory droplets or aerosols, and through the ingestion of viral particles. COVID-19 also affects multiple organs, often leading to organ failure and death of the individual affected by the infection. Organic complications observed during the infectious process include those seen in the lungs, cardiovascular and gastrointestinal systems, among others. All of these contribute to the enormous morbidity and mortality of SARS-CoV-2 infection.
References
- Sin-Yee Fung; Kit-San Yuen; Zi-Wei Ye; Chi-Ping Chan; Dong-Yan Jin; A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerging Microbes & Infections 2020, 9, 558-570, 10.1080/22221751.2020.1736644.
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J.; et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020, 69, 1010–1018.
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv 2020.
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020.
- B. Coutard; C. Valle; X. De Lamballerie; B. Canard; N.G. Seidah; E. Decroly; The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Research 2020, 176, 104742-104742, 10.1016/j.antiviral.2020.104742.
- Markus Hoffmann; Hannah Kleine-Weber; Simon Schroeder; Nadine Krüger; Tanja Herrler; Sandra Erichsen; Tobias S. Schiergens; Georg Herrler; Nai-Huei Wu; Andreas Nitsche; et al.Marcel A. MüllerChristian DrostenStefan Pöhlmann SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271-280.e8, 10.1016/j.cell.2020.02.052.
