The ENA ATPases (from exitus natru: the exit of sodium), also known as Na+-ATPases, are responsible for exchanges Na+ for H+; it belongs to the P-type ATPases are structurally very similar to the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA). ENA ATPases are essential for salt tolerance and in alkaline conditions, where other transporters cannot mediate an uphill Na+ efflux. P-type ATPases are classified in subclasses like types IIA, IIB, IIC, IID, and recently, was identified the type ATP4-type ATPases, related to Na+-ATPases found in Plasmodium falciparum and Toxoplasma gondii. In malaria, some drugs, like spiroindolones, targets PfATP4 and destroy Na+ homeostasis; these drugs are now in clinical trials. The ENA P-type (IID P-type ATPase) and ATP4-type ATPases have no structural homolog in mammalian cells, appearing only in fungi, plants, and protozoan parasites, e.g., Trypanosoma cruzi, Leishmania sp., Toxoplasma gondii, and Plasmodium falciparum. This exclusivity turns the Na+-ATPase a potential candidate for the biologically-based design of new therapeutic interventions; for this reason, Na+-ATPases deserves more attention.
- ENA P-type ATPase
- Type IID P-type ATPase
- ATP4-type ATPase
- Na+-ATPases
- Trypanosomatids
- Apicomplexa
- Spiroindolones
- Malaria clinical trials
1. Introduction
In the vast majority of cases, life has evolved in high-Na
+
environments [1], which are considered inadequate for, or injurious to, proper cellular functioning [2], notably without mechanisms that can maintain low intracellular Na
+
concentrations in organisms surrounded by and extracellular fluid containing ~150 mM Na
+
[3]. Thus, Na
+
extrusion from the intracellular milieu becomes a challenge and a prerequisite for the survival, growth, and the evolution of all species. Na
+
movement across the plasma membrane was initially described as being associated with K
+
movements. The first Na
+
pump described was (Na
+
+ K
+
)-ATPase. Two significant studies identified this pump: in the first, Skou [4] found that Na
+
and K
+
stimulated the catalysis of ATP hydrolysis by an ATPase in membrane fragments of leg nerves from the shore crab; in the second, Post & Jolly [5] demonstrated that Na
+
extrusion and K
+
uptake occur simultaneously in opposing directions and in an energy-dependent manner across the red-cell plasma membrane, with a stoichiometry of 3Na
+
:2K
+
.
A few years later, Proverbio et al. [6] described an ouabain-resistant Na
+
-ATPase activity that acts independently of K
+
in isolated membranes from the outermost cortex of a guinea pig, and that was sensitive. When they studied Na
+
fluxes through the basolateral membranes of proximal tubule cells towards the external milieu they found two mechanisms: (i) one coupled to K
+
and sensitive to ouabain, (ii) another one not coupled to K
+, resistant to ouabain and sensitive to ethacrynic acid [7]. More recently [8][9], the K
, resistant to ouabain and sensitive to ethacrynic acid [7]. More recently [8,9], the K
+
-independent and ouabain-resistant mechanism received the name of “second Na
+
pump”, which is still accepted [10].
The finding that the diuretic, furosemide, inhibits the ATP-dependent Na
+
efflux independently of K
+
(named for this reason Na
+
-ATPase) with no effect on the classic (Na
+
+ K
+
)-ATPase [11] was a significant step in identifying this Na
+
pump in several organisms. The main evolutionary advantage of a Na
+
-ATPase is its efficacy as a mechanism for Na
+
extrusion without interference with intracellular K
+ homeostasis. Rocafull et al. [9][12][13]purified and cloned the Na
homeostasis. Rocafull et al. [9,12,13] purified and cloned the Na
+
-ATPase from enterocytes (ATNA); its 3D structure was proposed, and the crucial amino acids of the catalytic and Na
+
binding sites were identified. They also demonstrated the enzyme’s sensitivity to furosemide and its resistance to ouabain. The existence of a second Na
+
pump—besides the (Na
+
+ K
+
)-ATPase—is now widely accepted.
The Na
+-ATPase from lower eukaryotes was first described in trypanosomatids [14][15][16]. In protozoan parasites, Na
-ATPase from lower eukaryotes was first described in trypanosomatids [14,15,16]. In protozoan parasites, Na
+
-ATPase, but not the (Na
+
+ K
+
)-ATPase, was found to be essential to energize the secondary active inorganic phosphate (P
i) transport [17][18]. In apicomplexan parasites, Na
) transport [17,18]. In apicomplexan parasites, Na
+-ATPase may be a therapeutic target for malaria and toxoplasmosis [19][20].
-ATPase may be a therapeutic target for malaria and toxoplasmosis [19,20].
2. Trypanosomatid Parasites
The first evidence of a Na
+
-ATPase in a trypanosomatid parasite was presented by Caruso-Neves et al. [14], who showed that ouabain, the specific (Na
+
+ K
+
)-ATPase inhibitor, did not completely abolish Na
+
-stimulated ATPase activity of
Trypanosoma cruzi
epimastigotes, suggesting that this parasite has a Na
+
-ATPase that is insensitive to ouabain. Iizumi et al. [15] confirmed this observation and identified the protein. The gene responsible for Na
+
-ATPase in
T. cruzi
was named TcENA, due to its similarity to the gene for Na
+-ATPase from plants and fungi called ENA (ENA from exitus natru: the exit of sodium) [21][22]. TcENA (
-ATPase from plants and fungi called ENA (ENA from exitus natru: the exit of sodium) [21,22]. TcENA (
) has 10 possible transmembrane domains (TMpred Server), as well as for highly conserved domains corresponding to the Type-II P-ATPases catalytic sites: (1) the signature sequence DKTGT
365
containing aspartic acid that is phosphorylated during the catalytic cycle; (2) the domain DGFND
700
involved in Mg
2+
binding (green circle), and (3) the conserved TGEA
203
sequence (red circle), as well as F
480
, K
485
and K
504 (orange circle), which are related to the nucleotide-binding domain. These conserved domains are related to the Type IID P-type group; therefore, their presence in T. cruzi allowed TcENA to be included in the Type IID P-ATPases group [20], i.e., within a family of ion-transporting ATPases that form a phosphorylated intermediate during the catalytic cycle [23][24][25]. Phylogenetic analysis allowed for the inclusion of TcENA within the unique group Type IID or ENA-type P-ATPases, which includes
(orange circle), which are related to the nucleotide-binding domain. These conserved domains are related to the Type IID P-type group; therefore, their presence in T. cruzi allowed TcENA to be included in the Type IID P-ATPases group [20], i.e., within a family of ion-transporting ATPases that form a phosphorylated intermediate during the catalytic cycle [23,24,25]. Phylogenetic analysis allowed for the inclusion of TcENA within the unique group Type IID or ENA-type P-ATPases, which includes
Leishmania braziliensis
and
L. donovani
,
Saccharomyces cerevisiae
, and
Entamoeba histolytica
[20].
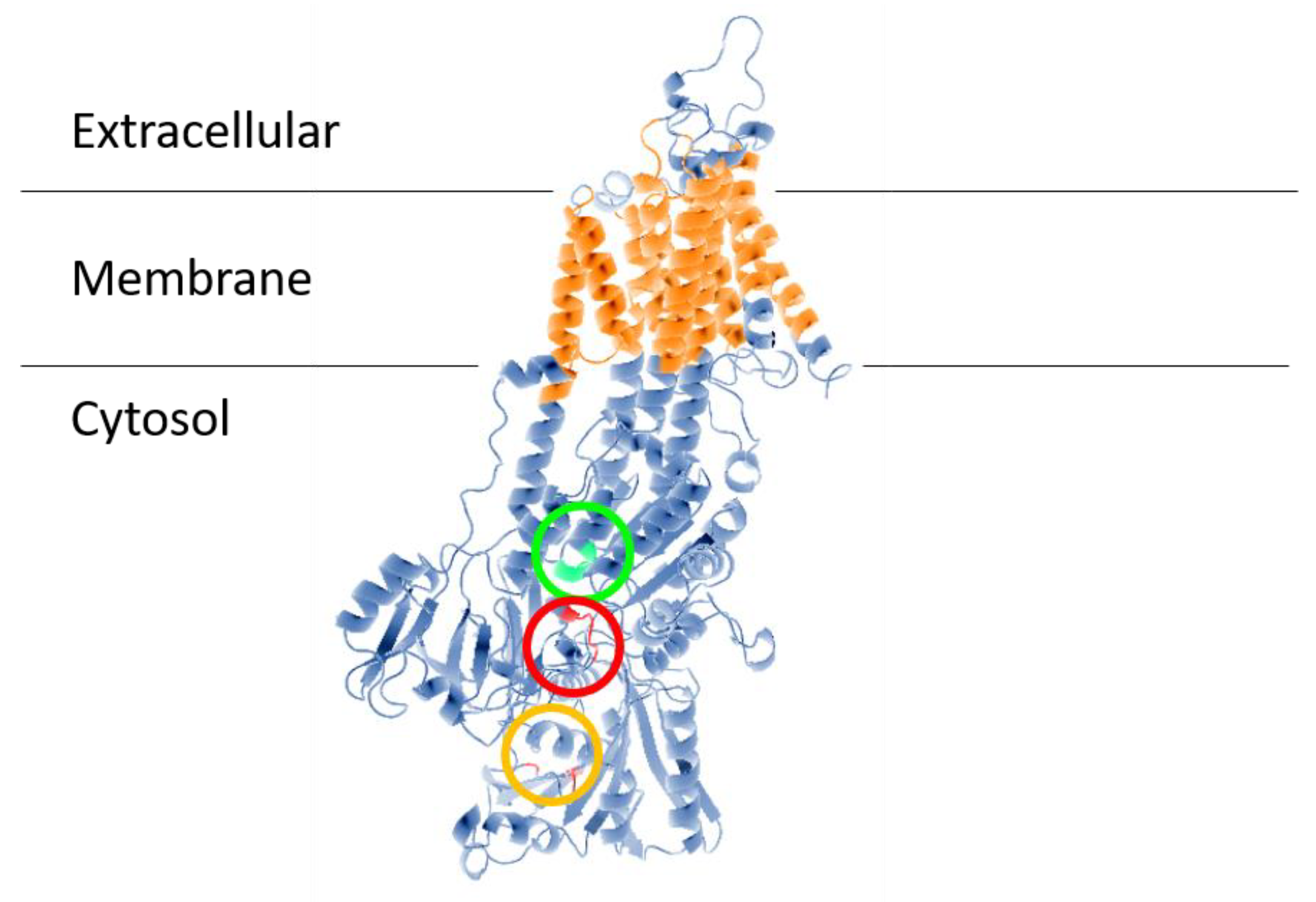
Figure 1.
Structural model of ENA ATPase from
T. cruzi
(TcENA). The model was constructed using the protein structure prediction PHYRE (
) [26], based on the model of the SERCA ATPase and visualized with the standard molecular viewer PyMOL 2002 (DeLano, W.L. The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA;
). The 10 transmembrane helices are shown in orange, and the key residues mentioned in the text are highlighted by a red circle (conserved TGEA domain), an orange circle (F
480
, K
485
, K
504
residues related to nucleotide-binding); and a green circle (DGFND
700
domain involved in Mg
2+
binding).
T. cruzi
, the etiological agent of Chagas’ Disease, is a parasitic protozoan with a complex life cycle involving morphologically and functionally different stages that enable these parasites to adapt to a variety of conditions. In insect vectors, the proliferative epimastigotes form is found in the midgut, while in mammalian hosts, the gut contains the nonproliferative trypomastigote and the proliferative amastigote forms [27]. It may be that at some stages of the protozoan life cycle, different parasites—not only
T. cruzi
—would have different sensitivity to drugs that target Na
+
-ATPase.
A critical role for ATPases is to contribute to the maintenance of the plasma membrane potential (ΔΨ), which is the result of asymmetrical charge distribution when concentration gradients of H
+
, Na
+
, K
+,
and Cl
−
are established in steady-state conditions. The ΔΨ of trypomastigotes forms is markedly sensitive to extracellular Na
+
and K
+
concentrations, and trypomastigotes have a high Na
+
-ATPase activity that contributes to a great extent in ΔΨ generation, in contrast with what happens in amastigotes. The Na
+
gradient in amastigote forms does not influence ΔΨ [28]. These observations match TcENA transcription levels, which vary among the different evolutionary forms of the parasite: trypomastigote and epimastigote forms have higher TcENA expression than amastigote forms [15]. Moreover, the subsequent addition of varying NaCl concentrations to a suspension of epimastigotes that were initially in Na
+
-free NMG medium (140 mM N-methylglucamine, 5 mM glucose, 1 mM MgSO
4
, 1 mM CaCl
2,
and 10 mM Hepes-Tris at pH 7.4) resulted in immediate and pronounced depolarization [28], consistent with the observation that Na
+
significantly stimulates ATPase activity of TcENA-expressing cells in a dose-dependent manner. Furthermore, epimastigotes overexpressing TcENA have salt tolerance, which is probably driven by their capacity for accelerated Na
+
efflux [15].