A critically ill polytrauma patient is one of the most complex cases to be admitted to the intensive care unit, due to both the primary traumatic complications and the secondary post-traumatic interactions. From a molecular, genetic, and epigenetic point of view, numerous biochemical interactions are responsible for the deterioration of the clinical status of a patient, and increased mortality rates. From a molecular viewpoint, microRNAs are one of the most complex macromolecular systems due to the numerous modular reactions and interactions that they are involved in. Regarding the expression and activity of microRNAs in sepsis, their usefulness has reached new levels of significance. MicroRNAs can be used both as an early biomarker for sepsis, and as a therapeutic target because of their ability to block the complex reactions involved in the initiation, maintenance, and augmentation of the clinical status.
Adapted from: Rogobete, A.F.; Sandesc, D.; Bedreag, O.H.; Papurica, M.; Popovici, S.E.; Bratu, T.; Popoiu, C.M.; Nitu, R.; Dragomir, T.; I. M. AAbed, H.; Ivan, M.V. MicroRNA Expression is Associated with Sepsis Disorders in Critically Ill Polytrauma Patients. Cells 2018, 7, 271. https://doi.org/10.3390/cells7120271
- sepsis
- microRNA
- inflammation
- biomarker
1. Biochemical and Biosynthesis Aspects of MicroRNAs
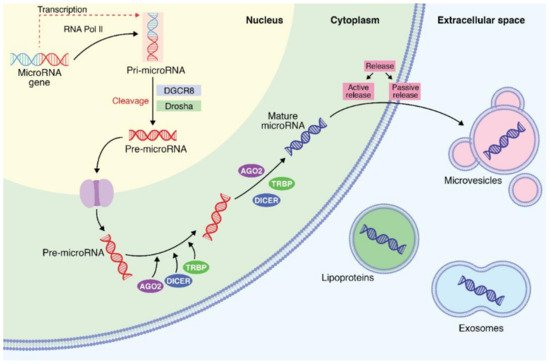
2. MicroRNA Identification from Different Body Fluids
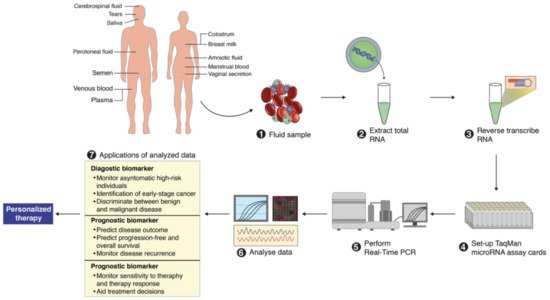
3. Importance of MicroRNAs for Clinical Use
4. Roles of MicroRNAs in the Pathophysiology of Oxidative Stress Associated with Sepsis
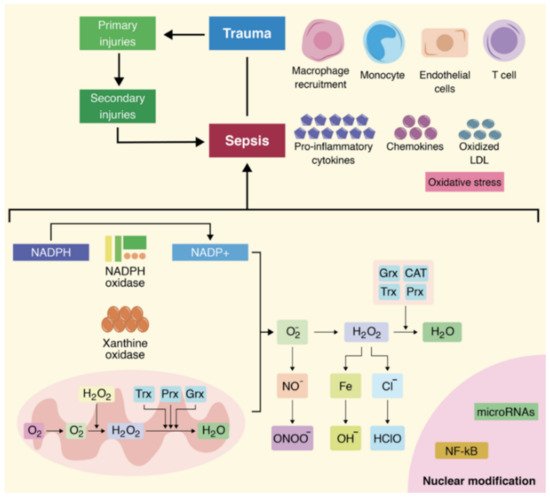
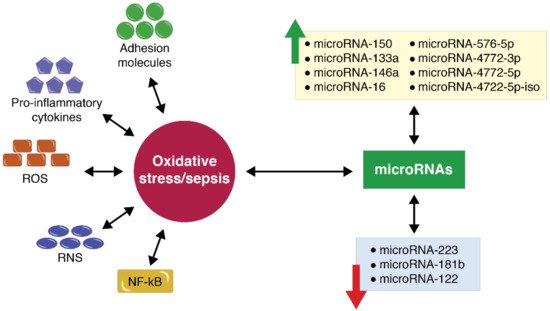
5. MicroRNA Expression in the Case of Polytrauma Patients with Sepsis
References
- David, L.V.; Ercisli, M.F.; Rogobete, A.F.; Boia, E.S.; Horhat, R.; Nitu, R.; Diaconu, M.M.; Pirtea, L.; Ciuca, I.; Horhat, D.; et al. Early prediction of sepsis incidence in critically Ill patients using specific genetic polymorphisms. Biochem. Genet. 2017, 55, 193–203.
- Essandoh, K.; Fan, G.C. Role of extracellular and intracellular microRNAs in sepsis. Biochim. Biophys. Acta 2014, 1842, 2155–2162.
- MacFarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561.
- Bratu, L.M.; Rogobete, A.F.; Papurica, M.; Sandesc, D.; Cradigati, C.A.; Sarandan, M.; Dumache, R.; Popovici, S.E.; Crisan, D.C.; Stanca, H.; et al. Literature research regarding miRNAs’ expression in the assessment and evaluation of the critically Ill polytrauma patient with traumatic brain and spinal cord injury. Clin. Lab. 2016, 62, 2019–2024.
- Macias, S.; Michlewski, G.; Cáceres, J.F. Hormonal regulation of microRNA biogenesis. Mol. Cell 2009, 36, 172–173.
- Bedreag, O.H.; Papurica, M.; Rogobete, A.F.; Sandesc, D.; Dumache, R.; Cradigati, C.A.; Sarandan, M.; Bratu, L.M.; Popovici, S.E.; Sima, L.V. Using circulating miRNAs as biomarkers for the evaluation and monitoring of the mitochondrial damage in the critically Ill polytrauma patients. Clin. Lab. 2016, 62, 1397–1403.
- Bedreag, O.H.; Sandesc, D.; Chiriac, S.D.; Rogobete, A.F.; Cradigati, A.C.; Sarandan, M.; Dumache, R.; Nartita, R.; Papurica, M. The use of circulating miRNAs as biomarkers for oxidative stress in critically Ill polytrauma patients. Clin. Lab. 2016, 62, 263–274.
- Bedreag, O.H.; Rogobete, A.F.; Sarandan, M.; Cradigati, A.C.; Papurica, M.; Dumbuleu, M.C.; Chira, A.M.; Rosu, O.M.; Sandesc, D. Oxidative stress in severe pulmonary trauma in critical ill patients. Antioxidant therapy in patients with multiple trauma—A review. Anaesthesiol. Intens. Ther. 2015, 47, 351–359.
- Ticlea, M.; Bratu, L.M.; Bodog, F.; Bedreag, O.H.; Rogobete, A.F.; Crainiceanu, Z.P. The use of exosomes as biomarkers for evaluating and monitoring critically Ill polytrauma patients with sepsis. Biochem. Genet. 2017, 55, 1–9.
- Sandesc, M.; Dinu, A.; Rogobete, A.F.; Bedreag, O.H.; Sandesc, D.; Papurica, M.; Bratu, L.M.; Negoita, S.; Vernic, C.; Popovici, S.E.; et al. Circulating microRNAs expressions as genetic biomarkers in pancreatic cancer patients continuous non-invasive monitoring. Clin. Lab. 2017, 63, 1561–1566.
- Negoita, S.I.; Sandesc, D.; Rogobete, A.F.; Dutu, M.; Bedreag, O.H.; Papurica, M.; Ercisli, M.F.; Popovici, S.E.; Dumache, R.; Sandesc, M.; et al. MiRNAs expressions and interaction with biological systems in patients with Alzheimer’s disease. Using miRNAs as a diagnosis and prognosis biomarker. Clin. Lab. 2017, 63, 1315–1321.
- Etheridge, A.; Lee, I.; Hood, L.; Galas, D.; Wang, K. Extracellular microRNA: A new source of biomarkers. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 717, 85–90.
- Guerra, R.C.; Zuñiga-muñoz, A.; Lans, V.G.; Díaz-díaz, E.; Alberto, C.; Betancourt, T.; Pérez-torres, I. Modulation of the activities of catalase, Cu-Zn, Mn superoxide dismutase, and glutathione peroxidase in adipocyte from ovariectomised female rats with metabolic syndrome. Int. J. Endocrinol. 2014, 2014.
- Arcaroli, J.J.; Hokanson, J.E.; Abraham, E.; Geraci, M.; Murphy, J.R.; Bowler, R.P.; Dinarello, C.A.; Silveira, L.; Sankoff, J.; Heyland, D.; et al. Extracellular superoxide dismutase haplotypes are associated with acute lung injury and mortality. Am. J. Respir. Crit. Care Med. 2009, 179, 105–112.
- Horhat, F.G.; Gundogdu, F.; David, L.V.; Boia, E.S.; Pirtea, L.; Horhat, R.; Cucui-Cozma, A.; Ciuca, I.; Diaconu, M.; Nitu, R.; et al. Early evaluation and monitoring of critical patients with acute respiratory distress syndrome (ARDS) using specific genetic polymorphisms. Biochem. Genet. 2017, 55, 204–211.
- Schmitt, B.; Vicenzi, M.; Garrel, C.; Denis, F.M. Redox biology effects of N-acetylcysteine, oral glutathione (GSH) and a novel sublingual form of GSH on oxidative stress markers: A comparative crossover study. Redox Biol. 2015, 6, 198–205.
- Hwang, J.H.; Ryu, J.; An, J.N.; Kim, C.T.; Kim, H.; Yang, J.; Ha, J.; Chae, D.W.; Ahn, C.; Jung, I.M.; et al. Pretransplant malnutrition, inflammation, and atherosclerosis affect cardiovascular outcomes after kidney transplantation. BMC Nephrol. 2015, 16, 109.
- Yin, G.; Wang, Y.; Cen, X.; Yang, M.; Liang, Y.; Xie, Q. Lipid peroxidation-mediated inflammation promotes cell apoptosis through activation of NF-kB pathway in rheumatoid arthritis synovial cells. Mediat. Inflamm. 2015, 2015, 460310.
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183.
- Tavladaki, T.; Spanaki, A.; Dimitriou, H.; Kozlov, A.; Duvigneau, J.; Weidinger, A.; Kondili, E.; Georgopoulos, D.; Briassoulis, G. Bioenergetics and metabolic patterns in early onset severe sepsis or trauma. Intensiv. Care Med. Exp. 2015, 3, A43.
- Denk, S.; Perl, M.; Huber-Lang, M. Damage- and pathogen-associated molecular patterns and alarmins: Keys to sepsis? Eur. Surg. Res. 2012, 48, 171–179.
- Soud, D.E.M.; Amin, O.A.I.; Amin, A.A.I. New era “soluble triggering receptor expressed on myeloid cells-I” as a marker for early detection of infection in trauma patients. Egypt. J. Anaesth. 2011, 27, 267–272.
- Horton, J.W. Free radicals and lipid peroxidation mediated injury in burn trauma: The role of antioxidant therapy. Toxicology 2003, 189, 75–88.
- Van der Kuip, M.; De Meer, K.; Oosterveld, M.J.S.; Lafeber, H.N.; Gemke, R.J.B.J. Simple and accurate assessment of energy expenditure in ventilated paediatric intensive care patients. Clin. Nutr. 2004, 23, 657–663.
- Andruszkow, H.; Fischer, J.; Sasse, M.; Brunnemer, U.; Andruszkow, J.H.K.; Gänsslen, A.; Hildebrand, F.; Frink, M. Interleukin-6 as inflammatory marker referring to multiple organ dysfunction syndrome in severely injured children. Scand. J. Trauma Resusc. Emerg. Med. 2014, 22, 16.
- Gu, W.; Jiang, J. Genetic polymorphisms and posttraumatic complications. Comp. Funct. Genom. 2010, 2010.
- Burkhardt, M.; Nienaber, U.; Pizanis, A.; Maegele, M.; Culemann, U.; Bouillon, B.; Flohé, S.; Pohlemann, T.; Paffrath, T. Acute management and outcome of multiple trauma patients with pelvic disruptions. Crit. Care 2012, 16, R163.
- Gao, J.; Zeng, L.; Zhang, A.; Wang, X.; Pan, W.; Du, D. Identification of haplotype tag single-nucleotide polymorphisms within the PPAR family genes and their clinical relevance in patients with major trauma. Int. J. Environ. Res. Public Health 2016, 13, 374.
- Zhao, C.; Sun, G.Q.; Ye, P.; Li, S.; Shia, Y. MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci. Rep. 2013, 3, 1329.
- Ramachandran, S.; Coffin, S.L.; Tang, T.Y.; Jobaliya, C.D.; Spengler, R.M.; Davidson, B.L. Cis-acting single nucleotide polymorphisms alter microRNA-mediated regulation of human brain-expressed transcripts. Hum. Mol. Genet. 2016, 25, 4939–4950.
- Yuva-Aydemir, Y.; Simkin, A.; Gascon, E.; Gao, F.B. MicroRNA-9—Functional evolution of a conserved small regulatory RNA. RNA Biol. 2011, 8, 557–564.
- Klinge, C.M. Estrogen regulation of microRNA expression. Curr. Genom. 2009, 10, 169–183.
- Fibach, E.; Dana, M. Oxidative stress in paroxysmal nocturnal hemoglobinuria and other conditions of complement-mediated hemolysis. Free Radic. Biol. Med. 2015, 88, 63–69.
- Abraham, E. Nuclear factor—kB and its role in sepsis-associated organ failure. J. Infect. Dis. 2003, 187, S364–S369.
- Bernardes, M.; Vieira, T.S.; Martins, M.J.; Lucas, R.; Costa, L.; Pereira, J.G.; Ventura, F.; Martins, E. Myocardial perfusion in rheumatoid arthritis patients: Associations with traditional risk factors and novel biomarkers. Biomed. Res. Int. 2017, 2017.
- Salsano, E.; Rizzo, A.; Bedini, G.; Bernard, L.; Olio, V.D.; Volorio, S.; Lazzaroni, M.; Ceccherini, I.; Lazarevic, D.; Cittaro, D.; et al. An autoinflammatory neurological disease due to interleukin 6 hypersecretion. J. Neuroinflamm. 2013, 10, 802.
- Treszl, A.; Kocsis, I.; Szathmári, M.; Schuler, Á.; Héninger, E.; Tulassay, T.; Vásárhelyi, B. Genetic variants of TNF-α, IL-1β, IL-4 receptor α-Chain, IL-6 and IL-10 genes are not risk factors for sepsis in low-birth-weight infants. Neonatology 2003, 83, 241–245.
- Harding, D.; Dhamrait, S.; Millar, A.; Humphries, S.; Marlow, N.; Whitelaw, A.; Montgomery, H. Is interleukin-6−174 genotype associated with the development of septicemia in preterm infants? Pediatrics 2003, 112, 800–803.
- Chuang, T.-Y.; Chang, H.-T.; Chung, K.-P.; Cheng, H.-S.; Liu, C.-Y.; Liu, Y.-C.; Huang, H.-H.; Chou, T.-C.; Chang, B.-L.; Lee, M.-R.; et al. High levels of serum macrophage migration inhibitory factor and interleukin 10 are associated with a rapidly fatal outcome in patients with severe sepsis. Int. J. Infect. Dis. 2014, 20, 13–17.
- Bedreag, O.H.; Rogobete, A.F.; Cradigati, C.A.; Sarandan, M.; Nartita, R.; Horhat, F.G.; Popovici, S.E.; Sandesc, D.; Papurica, M. A novel evaluation of microvascular damage in critically ill polytrauma patients by using circulating micrornas. Rev. Rom. Med. Lab. 2016, 24, 21–30.
- Scott, E.; Loya, K.; Mountford, J.; Milligan, G.; Baker, A.H. MicroRNA regulation of endothelial homeostasis and commitment—Implications for vascular regeneration strategies using stem cell therapies. Free Radic. Biol. Med. 2013, 64, 52–60.
- Hulsmans, M.; Holvoet, P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc. Res. 2013, 100, 7–18.
- Kung, C.; Hsiao, S.; Tsai, T.; Su, C.; Chang, W.; Huang, C.; Wang, H.; Lin, W.; Chang, H.; Lin, Y.; et al. Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J. Transl. Med. 2012, 10, 130.
- Haas, C. Lung protective mechanical ventilation in acute respiratory distress syndrome. Respir. Care Clin. 2003, 9, 363–396.
- Haas, S.; Trepte, C. Prediction of volume responsiveness using pleth variability index in patients undergoing cardiac surgery after cardiopulmonary bypass. J. Anesth. 2012, 26, 696–701.
- Wada, T.; Jesmin, S.; Gando, S.; Yanagida, Y.; Mizugaki, A.; Sultana, S.N.; Zaedi, S. The role of angiogenic factors and their soluble receptors in acute lung injury (ALI)/acute respiratory distress syndrome (ARDS) associated with critical illness. J. Inflamm. 2013, 10, 6.
- Medford, A.R.L.; Godinho, S.I.H.; Keen, L.J. Fluid vascular endothelial growth factor protein levels in patients with and at risk for ARDS. Chest 2009, 136, 457–464.
- Abadie, Y.; Bregeon, F.; Papazian, L.; Lange, F.; Thomas, P.; Duvaldestin, P.; Adnot, S. Decreased VEGF concentration in lung tissue and vascular injury during ARDS. Eur. Respir. J. 2005, 25, 139–146.
- Kondo, T.; Hattori, N.; Ishikawa, N.; Murai, H.; Haruta, Y.; Hirohashi, N.; Tanigawa, K.; Kohno, N. KL-6 concentration in pulmonary epithelial lining fluid is a useful prognostic indicator in patients with acute respiratory distress syndrome. Respir. Res. 2011, 12, 32.
- Aggarwal, N.R.; Alessio, F.R.D.; Tsushima, K.; Sidhaye, V.K.; Cheadle, C.; Grigoryev, D.N.; Barnes, K.C.; King, L.S. Regulatory T cell-mediated resolution of lung injury: Identification of potential target genes via expression profiling. Physiol. Genom. 2010, 41, 109–119.
- Pena, O.M.; Hancock, D.G.; Lyle, N.H.; Linder, A.; Russell, J.A.; Xia, J.; Fjell, C.D.; Boyd, J.H.; Hancock, R.E.W. An endotoxin tolerance signature predicts sepsis and organ dysfunction at initial clinical presentation. EBioMedicine 2014, 1, 64–71.
- McClure, C.; Brudecki, L.; Ferguson, D.A.; Yao, Z.Q.; Moorman, J.P.; McCall, C.E.; Gazzar, M. El microRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect. Immun. 2014, 82, 3816–3825.
- Belopolskaya, O.B.; Smelaya, T.V. Clinical associations of host genetic variations in the genes of cytokines in critically ill patients. Clin. Exp. Immunol. 2015, 180, 531–541.
- Jacobi, J. Pathophysiology of sepsis. Am. J. Heal Pharm. 2002, 59, 1435–1444.
- Puskarich, M.; Shapiro, N.; Trzeciak, S. Plasma levels of mitochondrial DNA in patients presenting to the emergency department with sepsis. Shock 2012, 38, 337–340.
- Fejes, Z.; Szilágyi, B.; Kappelmayer, J.; Ifj Nagy, B. Alteration in the expression of platelet microRNAs in diseases with abnormal platelet activation. Orv. Hetil. 2018, 159, 1962–1970.
- Hassan, F.I.; Didari, T.; Khan, F.; Mojtahedzadeh, M.; Abdollahi, M. The role of epigenetic alterations involved in sepsis: An overview. Curr. Pharm. Des. 2018, 24, 2862–2869.
- Ehrnthaller, C.; Flierl, M.; Perl, M.; Denk, S.; Unnewehr, H.; Ward, P.A.; Radermacher, P.; Ignatius, A.; Gebhard, F.; Chinnaiyan, A.; et al. The molecular fingerprint of lung inflammation after blunt chest trauma. Eur. J. Med. Res. 2015, 20, 70.
- Zhai, R.; Gong, M.N.; Zhou, W.; Thompson, T.B.; Kraft, P.; Su, L.; Christiani, D.C. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax 2007, 62, 718–722.
- Medford, A.R.; Millar, A.B. Vascular endothelial growth factor (VEGF) in acute lung injury (ALI) and acute respiratory distress syndrome (ARDS): Paradox or paradigm? Thorax 2006, 61, 621–626.
- Duan, Q.; Chen, C.; Yang, L.; Li, N.; Gong, W.; Li, S.; Wang, D.W. MicroRNA regulation of unfolded protein response transcription factor XBP1 in the progression of cardiac hypertrophy and heart failure in vivo. J. Transl. Med. 2015, 13, 363.
- Blass, S.C.; Goost, H.; Tolba, R.H.; Stoffel-Wagner, B.; Kabir, K.; Burger, C.; Stehle, P.; Ellinger, S. Time to wound closure in trauma patients with disorders in wound healing is shortened by supplements containing antioxidant micronutrients and glutamine: A. PRCT. Clin. Nutr. 2012, 31, 469–475.
- Yehya, N.; Yerrapureddy, A.; Tobias, J.; Margulies, S.S. MicroRNA modulate alveolar epithelial response to cyclic stretch. BMC Genom. 2012, 13, 154.
- Kulshreshtha, R.; Ferracin, M.; Wojcik, S.E.; Garzon, R.; Alder, H.; Agosto-Perez, F.J.; Davuluri, R.; Liu, C.-G.; Croce, C.M.; Negrini, M.; et al. A microRNA signature of hypoxia. Mol. Cell Biol. 2007, 27, 1859–1867.
- Moschos, S.A.; Williams, A.E.; Perry, M.M.; Birrell, M.A.; Belvisi, M.G.; Lindsay, M.A. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genom. 2007, 8, 240.
- Tacke, F.; Roderburg, C.; Benz, F.; Cardenas, D.V.; Luedde, M.; Hippe, H.-J.; Frey, N.; Vucur, M.; Gautheron, J.; Koch, A.; et al. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically Ill patients. Crit. Care Med. 2014, 42, 1096–1104.
- Wang, X.; Huang, W.; Yang, Y.; Wang, Y.; Peng, T.; Chang, J.; Caldwell, C.C.; Zingarelli, B.; Fan, G. Biochimica et biophysica acta loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. BBA Mol. Basis Dis. 2014, 1842, 701–711.
- Shao, Y.; Li, J.; Cai, Y.; Xie, Y.; Ma, G.; Li, Y.; Chen, Y.; Liu, G.; Zhao, B.; Cui, L.; et al. The functional polymorphisms of miR-146a are associated with susceptibility to severe sepsis in the Chinese population. Mediat. Inflamm. 2014, 2014.
- Søndergaard, E.S.; Alamili, M.; Coskun, M.; Gögenur, I. MicroRNA’s are novel biomarkers in sepsis—A systematic review. Trends Anaesth. Crit. Care 2015, 5, 151–156.
- Roderburg, C.; Luedde, M.; Vargas Cardenas, D.; Vucur, M.; Scholten, D.; Frey, N.; Koch, A.; Trautwein, C.; Tacke, F.; Luedde, T. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS ONE 2013, 8, e54612.
- La Rosa, A.H.; Acker, M.; Swain, S.; Manoharan, M. The role of epigenetics in kidney malignancies. Cent European. J. Urol. 2015, 68, 157–164.
- Xu, S.; Zhang, R.; Niu, J.; Cui, D.; Xie, B.; Zhang, B. Oxidative stress mediated-alterations of the microRNA expression profile in mouse hippocampal neurons. Int. J. Mol. Sci. 2012, 13, 16945–16960.
- Yao, L.; Liu, Z.; Zhu, J.; Li, B.; Chai, C.; Tian, Y. Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. Int. J. Clin. Exp. Pathol. 2015, 8, 7675–7684.
- Rodrigues, C.E.; Capcha, J.M.C.; De Bragança, A.C.; Sanches, T.R.; Gouveia, P.Q.; De Oliveira, P.A.F.; Malheiros, D.M.A.C.; Volpini, R.A.; Santinho, M.A.R.; Santana, B.A.A.; et al. Human umbilical cord-derived mesenchymal stromal cells protect against premature renal senescence resulting from oxidative stress in rats with acute kidney injury. Stem Cell Res. Ther. 2017, 8, 19.
- Fredriksson, K.; Tjäder, I.; Keller, P.; Petrovic, N.; Ahlman, B.; Schéele, C.; Wernerman, J.; Timmons, J.A.; Rooyackers, O. Dysregulation of mitochondrial dynamics and the muscle transcriptome in ICU patients suffering from sepsis induced multiple organ failure. PLoS ONE 2008, 11, e3686.
- Huang, C.; Xiao, X.; Chintagari, N.R.; Breshears, M.; Wang, Y.; Liu, L. MicroRNA and mRNA expression profiling in rat acute respiratory distress syndrome. BMC Med. Genom. 2014, 7, 46.
- Bedreag, O.H.; Rogobete, A.F.; Dumache, R.; Sarandan, M.; Cradigati, A.C.; Papurica, M.; Craciunescu, M.C.; Popa, D.M.; Luca, L.; Nartita, R.; et al. Use of circulating microRNAs as biomarkers in critically ill polytrauma patients. Biomarkers Genom. Med. 2015, 7, 131–138.
- Benz, F.; Roy, S.; Traukwein, C.; Roderburg, C.; Luedde, T. Circulating microRNAs as biomarkers for sepsis. Int. J. Mol. Sci. 2016, 17, 78.
- Huang, J.; Sun, Z.; Yan, W.; Zhu, Y.; Lin, Y.; Chen, J.; Shen, B.; Wang, J. Identfifcation of microRNAs as sepsis biomarkers based on MiRNA regulatory network analysis. Biomed. Res. Int. 2014, 2014, 594350.
- Dumache, R.; Rogobete, A.F.; Bedreag, O.H.; Sarandan, M.; Cradigati, A.C.; Papurica, M.; Dumbuleu, C.M.; Nartita, R.; Sandesc, D. Use of miRNAs as biomarkers in sepsis. Anal. Cell. Pathol. 2015, 2015, 186716.
- Sun, Z.; Zhang, Q.; Cui, X.; Yang, J.; Zhang, B.; Song, G. Differential expression of miRNA and its role in sepsis. Pediatrics 2018, 142.
- Ansari, M.; Gupta, P. Nex-Gen biomarkers—A genetic model of sepsis. Biomark. J. 2016, 2, 8.
- Vasilescu, C.; Dragomir, M.; Tanase, M.; Giza, D.; Purnichescu-Purtan, R.; Chen, M.; Yeung, S.C.-J.; Calin, G.A. Circulating miRNAs in sepsis—A network under attack: An in silico perdiction of the potential existance of microRNAs sponges in sepsis. PLoS ONE 2017, 12, e01833334.
- Puskarich, M.A.; Nandi, U.; Shapiro, N.I.; Trzeciak, S.; Kline, J.A.; Jones, A.E. Detection of microRNAs in patients with sepsis. J. Acute Dis. 2015, 4, 101–106.
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741.
- Díaz-Prado, S.; Cicione, C.; Muiños-López, E.; Hermida-Gómez, T.; Oreiro, N.; Fernández-López, C.; Blanco, F.J. Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC Musculoskelet. Disord. 2012, 13, 144.
- Campomenosi, P.; Gini, E.; Noonan, D.M.; Poli, A.; D’Antona, P.; Rotolo, N.; Dominioni, L.; Imperatori, A. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol. 2016, 16, 60.
- Warburton, A.; Savage, A.L.; Myers, P.; Peeney, D.; Bubb, V.J.; Quinn, J.P. Molecular signatures of mood stabilisers highlight the role of the transcription factor REST/NRSF. J. Affect. Disord. 2014, 172, 63–73.
- Inchley, C.S.; Sonerud, T.; Fjærli, H.O.; Nakstad, B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect. Dis. 2015, 15, 150.
- Dong, J.; Liu, Y.; Liao, W.; Liu, R.; Shi, P.; Wang, L. MiRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J. Bone Oncol. 2016, 5, 74–79.
- Hu, Z.; Chen, X.; Zhao, Y.; Tian, T.; Jin, G.; Shu, Y.; Chen, Y.; Xu, L.; Zen, K.; Zhang, C.; et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010, 28, 1721–1726.
- Pandey, A.C.; Semon, J.A.; Kaushal, D.; O’Sullivan, R.P.; Glowacki, J.; Gimble, J.M.; Bunnell, B.A. MicroRNA profiling reveals age-dependent differential expression of nuclear factor kappaB and mitogen-activated protein kinase in adipose and bone marrow-derived human mesenchymal stem cells 928. Stem Cell Res. Ther. 2011, 2, 49.
- Haider, B.A.; Baras, A.S.; McCall, M.N.; Hertel, J.A.; Cornish, T.C.; Halushka, M.K. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS ONE 2014, 9.
- Zheng, D.; Yu, Y.; Li, M.; Wang, G.; Chen, R.; Fan, G.C.; Martin, C.; Xiong, S.; Peng, T. Inhibition of microRNA 195 prevents apoptosis and multiple-organ injury in mouse models of sepsis. J. Infect. Dis. 2016, 213, 1661–1670.
- Fang, C.; Zhao, J.; Li, J.; Qian, J.; Liu, X.; Sun, Q.; Liu, W.; Tian, Y.; Ji, A.; Wu, H.; et al. Massively parallel sequencing of microRNA in bloodstains and evaluation of environmental influences on miRNA candidates using realtime polymerase chain reaction. Forensic. Sci. Int. Genet. 2018, 38, 32–38.
- Tian, F.; Yuan, C.; Hu, L.; Shan, S. MicroRNA-93 inhibits inflammatory responses and cell apoptosis after cerebral ischemia reperfusion by targeting interleukin-1 receptor-associated kinase 4. Exp. Ther. Med. 2017, 14, 2903–2910.
- Qiu, J.; Zhou, X.; Zhou, X.; Cheng, R.; Liu, H.; Li, Y. Neuroprotective effects of microRNA-210 on hypoxic-ischemic encephalopathy. BioMed Res. Int. 2013, 2013, 350419.
- Hara, N.; Kikuchi, M.; Miyashita, A.; Hatsuta, H.; Saito, Y.; Kasuga, K.; Murayama, S.; Ikeuchi, T.; Kuwano, R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017, 5, 10.
- Xiao, B.; Liu, H.; Gu, Z. Expression of microRNA-133 inhibits epithelial—Mesenchymal transition in lung cancer cells by directly targeting FOXQ1. Arch. Bronconeumol. 2016, 52, 505–511.
- Liz, J.; Esteller, M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 169–176.
- Bera, A.; Ghosh-Choudhury, N.; Dey, N.; Das, F.; Kasinath, B.S.; Abboud, H.E.; Ghosh, G. NF-κB-mediated cyclin D1 expression by microRNA-21 in fluences renal cancer cell proliferation. Cell Signal. 2013, 25, 2575–2586.
- Huang, J.; Liu, J.; Chen-Xiao, K.; Zhang, X.; Lee, W.N.P.; Go, V.L.W.; Xiao, G.G. Advance in microRNA as a potential biomarker for early detection of pancreatic cancer. Biomark. Res. 2016, 4, 20.
- Li, S.; Liang, Z.; Xu, L.; Zou, F. MicroRNA-21: A ubiquitously expressed pro-survival factor in cancer and other diseases. Mol. Cell. Biochem. 2012, 360, 147–158.
- Zhou, X.; Su, S.; Li, S.; Pang, X.; Chen, C.; Li, J. MicroRNA-146a down-regulation correlates with neuroprotection and targets pro-apoptotic genes in cerebral ischemic injury in vitro. Brain Res. 2016, 1648, 136–143.
- Dumache, R.; Ciocan, V.; Muresan, C.; Enache, A. Molecular DNA analysis in forensic identification. Clin. Lab. 2016, 62, 245–248.
- Horhat, F.G.; Rogobete, A.F.; Papurica, M.; Sandesc, D.; Tanasescu, S.; Dumitrascu, V.; Licker, M.; Nitu, R.; Cradigati, C.A.; Sarandan, M.; et al. The use of lipid peroxidation expression as a biomarker for the molecular damage in the critically Ill polytrauma patient. Clin. Lab. 2016, 62, 1601–1607.
- Papurica, M.; Rogobete, A.F.; Sandesc, D.; Cradigati, C.A.; Sarandan, M.; Crisan, D.C.; Horhat, F.G.; Boruga, O.; Dumache, R.; Nilima, K.R.; et al. The expression of nuclear transcription factor kappa B (NF-κB) in the case of critically Ill polytrauma patients with sepsis and its interactions with microRNAs. Biochem. Genet. 2016, 54, 337–347.
