Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Cristina Cristofoletti.
Sézary syndrome (SS) is an aggressive variant of cutaneous t-cell lymphoma characterized by the accumulation of neoplastic CD4+ lymphocytes—the SS cells—mainly in blood, lymph nodes, and skin. The tumor spread pattern of SS makes this lymphoma a unique model of disease that allows a concurrent blood and skin sampling for analysis.
- cutaneous T-cell lymphoma
- Sézary syndrome
1. Introduction
Cutaneous T-cell lymphomas (CTCLs) include a large spectrum of mature T-cell neoplasms characterized by the accumulation of neoplastic CD4+ T lymphocytes in the skin.
The most common subtype of CTCL is mycosis fungoides (MF), which represents 60% of CTCL cases, while the much rarer variant is Sézary syndrome (SS), which accounts for approximately 5% [1]. The majority of patients with an early MF show a skin-restricted infiltration of malignant cells and an indolent course [1], but about 15% of them can progress toward advanced stages characterized by tumors and erythroderma and that show a decreased survival at less than 5 years [2,3][2][3]. On the contrary, SS is a rare and leukemic subtype of CTCL showing, ab initio, the simultaneous presence of neoplastic lymphocytes mainly in the blood, lymph nodes, and skin.
The relationship between MF and SS is still debated. For a long time, SS was considered an aggressive form of MF, but growing data from genome analyses have demonstrated specific chromosomal alterations occurring in MF but not in SS, and vice versa, suggesting that they are two separate clinical entities [4]. These findings are also reinforced by the different transcriptomic profiles and cell surface markers expressed by MF and SS. According to these data, Campbell et al. indicated that MFs arise from skin resident effector memory (EM) T cells, whereas SS arises from central memory (CM) T-lymphocytes [5]. However, the cell of origin of SS and MF is still undetermined.
Despite the great efforts to characterize SS pathogenesis, it remains an incurable disease, with 5-year overall survival (OS) rates of 15% to 40% for stage IVA and of 0% to 15% for stage IVB [2,6][2][6]. To date, most studies have explored the intrinsic molecular features of SS, demonstrating that it is characterized by a complex profile of chromosome aberrations [7,8,9][7][8][9] and a broad range of genes variously affected by somatic copy-number alterations and somatic single-nucleotide variants that are involved predominantly in T-cell activation and apoptosis, activation of NF-kB, JAK/STAT signaling, chromatin remodeling, and DNA damage response [10,11][10][11].
More recently, attention has shifted toward extrinsic mechanisms that impact the pathophysiology of cancer. Indeed, understanding how the tumor microenvironment sustains neoplastic cells has become focal in cancer research [12]. Regarding CTCL, many reports have already described the complex cellular interactions between malignant lymphocytes and skin elements that play a key role in CTCL pathogenesis [13,14,15,16,17][13][14][15][16][17].
Unlike metastatic spread, cutaneous lymphoma dissemination does not reflect tumor progression, but rather a conserved physiological behavior of neoplastic lymphocytes [18]. This characteristic indicates the strong dependence of these cells on the cutaneous niche that supports their proliferation and survival through the release of nutrients and the induction of cellular signals [19,20,21,22,23][19][20][21][22][23]. Tumor is a dynamic disease, and during its progression usually acquires a greater heterogeneity. This implies that the original cancer cells acquire different molecular signatures, generating an intratumor heterogeneity. Such a condition is the greatest cause of failure in cancer therapies because the spatial and temporal genetic and epigenetic diversity and/or plastic gene expression in cancer cells is often associated with the mechanism of drug resistance [24]. A strong contribution to heterogeneity is also made by the tumor microenvironment, which triggers distinct and specific signals to tumor cells. This last aspect assumes a particular importance in CTCL, given the SS cells simultaneously invade the bloodstream and the skin district, where they interact with keratinocytes, as well as bystander stromal and immune cells [25]. The impact of fluid and solid environment on tumor cells adds complexity to the disease and produces intratumor heterogeneity.
2. Blood and Skin Microenvironments Drive the Transcriptional Program and Signaling of SS Cells
A role for the skin microenvironment in SS pathogenesis has long been hypothesized, but its function in vivo is poorly known. Lately, some investigations have analyzed matched blood- and skin-derived SS cells, revealing substantial differences between these two cell subsets. In this section, we discuss the main results obtained so far will be discussed (Figure 1).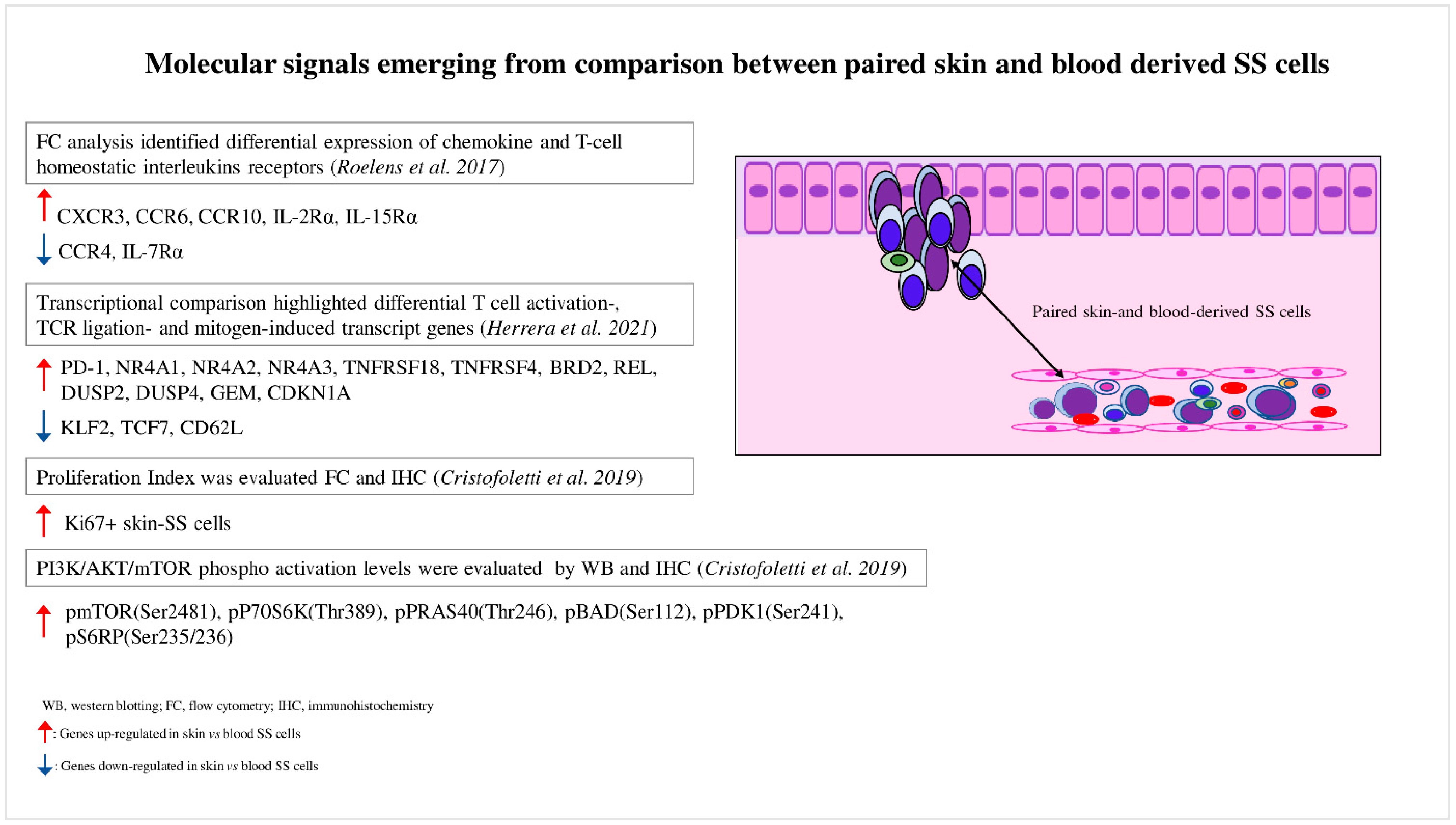
Figure 1. Molecular signals emerging from comparison between paired skin- and blood-derived SS cells. Comparison analyses performed between paired skin- and blood-derived SS cells highlight the molecular signals triggered in these two SS cell subpopulations.
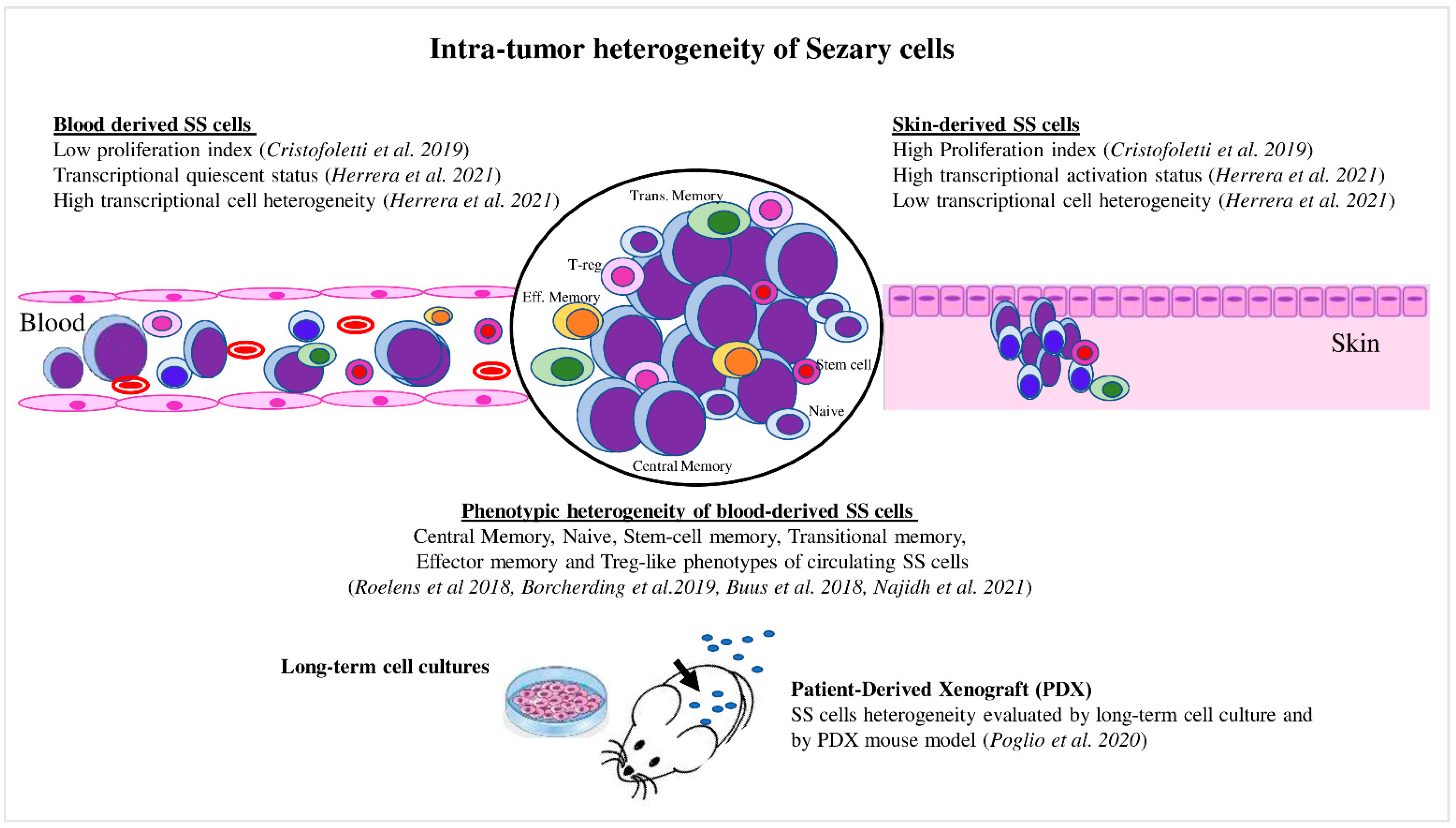
Figure 2. Intratumor heterogeneity of Sézary cells. Skin- and blood-derived SS cells showed differences in proliferation capacity, expression of activation markers, and level of transcriptional heterogeneity. Blood SS cells showed phenotypic heterogeneity, which also was evaluated under specific culture conditions and in the PDX mouse model.
3. Skin SS Cells Exhibit a Hyperactivated PI3K/AKT/mTOR Signaling Compared to the Matched Blood Counterpart: Spotlight on Skin Microenvironment
Activation of CD4+ T cells upon engagement of the TCR and costimulatory receptors lead to changes in expression profiles, remodeling of the T-cell proteome, and differentiation into effector CD4+ T-cell subpopulations [36][34]. TCR pathway alterations are frequently observed in SS [7,9,37,38,39,40][7][9][35][36][37][38]. Engagement of TCR activates, among others, the PI3K/AKT/mTOR pathway, leading to T-cell proliferation, survival, and differentiation, as well as cytokine production [41][39]. Throughout the years, many investigations proved that PI3K/AKT/mTOR signaling is strongly involved in SS pathogenesis. Early genomic studies demonstrated that PTEN, the major antagonist of PI3K/AKT signaling [42][40], is frequently deleted at the monoallelic level in both MF [43][41] and SS, in which PTEN is also downregulated by miR-21, miR-106b, and miR-486 [44][42]. More recently, oura study investigated the CNVs of members belonging to this cascade in 43 SS patients highlighting recurrent alterations; namely, a loss in tumor suppressors such as LKB1 (48%), PTEN (39%), and PDCD4 (35%), and a gain in the proto-oncogene P70S6K (30%). Each of these CNVs, whether evaluated individually or in combination, was associated with the reduced survival of SS patients [31][29]. It was notable that these genetic alterations were differently distributed among patients, thus contributing to the interpatient heterogeneity. According to these results, the therapeutic efficacy of PI3K/mTOR inhibitors was found to be variable among patients and correlated with their genomic status, thus confirming the strong impact of heterogeneity in the clinical course of SS patients as well [45][43]. Functional experiments demonstrated that many cytokines and growth factors detected in SS skin lesions are able to activate PI3K/AKT/mTOR signaling. The first evidence came from Marzec M. et al. [46][44], who demonstrated that IL-2 triggered this pathway in activated primary SS cells and that its inhibition, obtained via treatment with the mTORC1 inhibitor rapamycin [46[44][45],47], was able to block SS cell growth in vitro [46][44] as well in a xenograft T-cell lymphoma mouse model [48][46]. Other investigations have been performed in this direction: Murga-Zamalloa et al. [49][47] studied the interaction between CTCL cells and lymphoma-associated macrophages known to play a critical role in disease progression, and demonstrated that autocrine colony stimulating factor 1 (CSF1) activated AKT/mTOR signaling and promoted T-cell lymphoma viability. Activation occurred upon binding to the CSF1 receptor, which is highly expressed by both macrophages and lymphoma cells. It is also interesting that the IL-31/IL-31 receptor axis involved in the mechanism of itch, which is one of the worst symptoms of SS [50][48], is capable of driving PI3K/AKT signaling in a plethora of skin diseases [51][49]. Immunohistochemistry confirmed that this pathway is activated in skin-infiltrating SS cells by the detection of high levels of phosphorylated forms of AKT, mTOR, P70S6K, S6RP, and 4EBP1 [46,52][44][50]. a Weresearch further deepened this aspect [31][29] by demonstrating a higher phosphorylation level of these proteins, particularly of mTOR, as well as a greater proliferation index in skin-derived SS cells when compared to blood-derived SS cells concurrently obtained from the same patients (Figure 1). We also proved that SDF-1 and CCL21 chemokines were also proved, both overexpressed in SS tissues [53,54,55,56][51][52][53][54], induced mTORC1 signaling activation, cell proliferation, and Ki67 upregulation in primary-SS cells and in a SS-derived cell line. The skin–blood comparison approach therefore demonstrates how the skin upregulates PI3K/AKT/mTORC1 signaling, and indicates a strategy for discovering new biochemical signals that support the growth and expansion of potentially compound target SS cells (Figure 1).References
- Scarisbrick, J.J.; Quaglino, P.; Prince, H.M.; Papadavid, E.; Hodak, E.; Bagot, M.; Servitje, O.; Berti, E.; Ortiz-Romero, P.; Stadler, R.; et al. The PROCLIPI International Registry of Early-Stage Mycosis Fungoides Identifies Substantial Diagnostic Delay in Most Patients. Br. J. Dermatol. 2019, 181, 350–357.
- Scarisbrick, J.J.; Prince, H.M.; Vermeer, M.H.; Quaglino, P.; Horwitz, S.; Porcu, P.; Stadler, R.; Wood, G.S.; Beylot-Barry, M.; Pham-Ledard, A.; et al. Cutaneous Lymphoma International Consortium Study of Outcome in Advanced Stages of Mycosis Fungoides and Sézary Syndrome: Effect of Specific Prognostic Markers on Survival and Development of a Prognostic Model. J. Clin. Oncol. 2015, 33, 3766–3773.
- Quaglino, P.; Pimpinelli, N.; Berti, E.; Calzavara-Pinton, P.; Alfonso Lombardo, G.; Rupoli, S.; Alaibac, M.; Bottoni, U.; Carbone, A.; Fava, P.; et al. Time Course, Clinical Pathways, and Long-Term Hazards Risk Trends of Disease Progression in Patients with Classic Mycosis Fungoides: A Multicenter, Retrospective Follow-up Study from the Italian Group of Cutaneous Lymphomas. Cancer 2012, 118, 5830–5839.
- Jonak, C.; Tittes, J.; Brunner, P.M.; Guenova, E. Mycosis Fungoides Und Sézary-Syndrom. JDDG J. der Dtsch. Dermatologischen Gesellschaft 2021, 19, 1307–1335.
- Campbell, J.J.; Clark, R.A.; Watanabe, R.; Kupper, T.S. Sezary Syndrome and Mycosis Fungoides Arise from Distinct T-Cell Subsets: A Biologic Rationale for Their Distinct Clinical Behaviors. Blood 2010, 116, 767–771.
- Olsen, E.; Vonderheid, E.; Pimpinelli, N.; Willemze, R.; Kim, Y.; Knobler, R.; Zackheim, H.; Duvic, M.; Estrach, T.; Lamberg, S.; et al. Revisions to the Staging and Classification of Mycosis Fungoides and Sézary Syndrome: A Proposal of the International Society for Cutaneous Lymphomas (ISCL) and the Cutaneous Lymphoma Task Force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007, 110, 1713–1722.
- Wang, L.; Ni, X.; Covington, K.R.; Yang, B.Y.; Shiu, J.; Zhang, X.; Xi, L.; Meng, Q.; Langridge, T.; Drummond, J.; et al. Genomic Profiling of Sézary Syndrome Identifies Alterations of Key T Cell Signaling and Differentiation Genes. Nat. Genet. 2015, 47, 1426–1434.
- Vermeer, M.H.; Van Doorn, R.; Dijkman, R.; Mao, X.; Whittaker, S.; Van Voorst Vader, P.C.; Gerritsen, M.J.P.; Geerts, M.L.; Gellrich, S.; Söderberg, O.; et al. Novel and Highly Recurrent Chromosomal Alterations in Sézary Syndrome. Cancer Res. 2008, 68, 2689–2698.
- Da Silva Almeida, A.C.; Abate, F.; Khiabanian, H.; Martinez-Escala, E.; Guitart, J.; Tensen, C.P.; Vermeer, M.H.; Rabadan, R.; Ferrando, A.; Palomero, T. The Mutational Landscape of Cutaneous T Cell Lymphoma and Sézary Syndrome. Nat. Genet. 2015, 47, 1465–1470.
- Tensen, C.P.; Quint, K.D.; Vermeer, M.H. Genetic and Epigenetic Insights into Cutaneous T-Cell Lymphoma. Blood 2022, 139, 15–33.
- Mirza, A.S.; Horna, P.; Teer, J.K.; Song, J.; Akabari, R.; Hussaini, M.; Sokol, L. New Insights Into the Complex Mutational Landscape of Sézary Syndrome. Front. Oncol. 2020, 10, 514.
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15.
- Miyagaki, T.; Sugaya, M. Immunological Milieu in Mycosis Fungoides and Sézary Syndrome. J. Dermatol. 2014, 41, 11–18.
- Krejsgaard, T.; Lindahl, L.M.; Mongan, N.P.; Wasik, M.A.; Litvinov, I.V.; Iversen, L.; Langhoff, E.; Woetmann, A.; Odum, N. Malignant Inflammation in Cutaneous T-cell Lymphoma—A Hostile Takeover. Semin. Immunopathol. 2017, 39, 269–282.
- Pileri, A.; Guglielmo, A.; Fuligni, F.; Lastrucci, I.; Patrizi, A.; Pimpinelli, N. Second Neoplasm in Cutaneous T-Cell Lymphoma Patients: A Marker of Worse Prognosis? Ital. J. Dermatology Venereol. 2021, 156, 484–488.
- Quaglino, P.; Fava, P.; Pileri, A.; Grandi, V.; Sanlorenzo, M.; Panasiti, V.; Guglielmo, A.; Alberti-Violetti, S.; Novelli, M.; Astrua, C.; et al. Phenotypical Markers, Molecular Mutations, and Immune Microenvironment as Targets for New Treatments in Patients with Mycosis Fungoides and/or Sézary Syndrome. J. Investig. Dermatol. 2021, 141, 484–495.
- Gonzalez, B.R.; Zain, J.; Rosen, S.T.; Querfeld, C. Tumor Microenvironment in Mycosis Fungoides and Sézary Syndrome. Curr. Opin. Oncol. 2016, 28, 88–96.
- Pals, S.T.; De Gorter, D.J.J.; Spaargaren, M. Lymphoma Dissemination: The Other Face of Lymphocyte Homing. Blood 2007, 110, 3102–3111.
- Yamanaka, K.; Clark, R.; Rich, B.; Dowgiert, R.; Hirahara, K.; Hurwitz, D.; Shibata, M.; Mirchandani, N.; Jones, D.A.; Goddard, D.S.; et al. Skin-Derived Interleukin-7 Contributes to the Proliferation of Lymphocytes in Cutaneous T-Cell Lymphoma. Blood 2006, 107, 2440–2445.
- Berger, C.L.; Hanlon, D.; Kanada, D.; Dhodapkar, M.; Lombillo, V.; Wang, N.; Christensen, I.; Howe, G.; Crouch, J.; El-Fishawy, P.; et al. The Growth of Cutaneous T-Cell Lymphoma Is Stimulated by Immature Dendritic Cells. Blood 2002, 99, 2929–2939.
- Wilcox, R.A.; Wada, D.A.; Ziesmer, S.C.; Elsawa, S.F.; Comfere, N.I.; Dietz, A.B.; Novak, A.J.; Witzig, T.E.; Feldman, A.L.; Pittelkow, M.R.; et al. Monocytes Promote Tumor Cell Survival in T-Cell Lymphoproliferative Disorders and Are Impaired in Their Ability to Differentiate into Mature Dendritic Cells. Blood 2009, 114, 2936–2944.
- Wu, X.; Schulte, B.C.; Zhou, Y.; Haribhai, D.; Mackinnon, A.C.; Plaza, J.A.; Williams, C.B.; Hwang, S.T. Depletion of M2-like Tumor-Associated Macrophages Delays Cutaneous T-Cell Lymphoma Development in Vivo. J. Investig. Dermatol. 2014, 134, 2814–2822.
- Rabenhorst, A.; Schlaak, M.; Heukamp, L.C.; Förster, A.; Theurich, S.; von Bergwelt-Baildon, M.; Büttner, R.; Kurschat, P.; Mauch, C.; Roers, A.; et al. Mast Cells Play a Protumorigenic Role in Primary Cutaneous Lymphoma. Blood 2012, 120, 2042–2054.
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94.
- Stolearenco, V.; Namini, M.R.J.; Hasselager, S.S.; Gluud, M.; Buus, T.B.; Willerslev-Olsen, A.; Ødum, N.; Krejsgaard, T. Cellular Interactions and Inflammation in the Pathogenesis of Cutaneous T-Cell Lymphoma. Front. Cell Dev. Biol. 2020, 8, 851.
- Roelens, M.; Delord, M.; Ram-Wolff, C.; Marie-Cardine, A.; Alberdi, A.; Maki, G.; Homyrda, L.; Bensussan, A.; Bagot, M.; Toubert, A.; et al. Circulating and Skin-Derived Sézary Cells: Clonal but with Phenotypic Plasticity. Blood 2017, 130, 1468–1471.
- Kim, Y.H.; Bagot, M.; Pinter-Brown, L.; Rook, A.H.; Porcu, P.; Horwitz, S.M.; Whittaker, S.; Tokura, Y.; Vermeer, M.; Zinzani, P.L.; et al. Mogamulizumab versus Vorinostat in Previously Treated Cutaneous T-Cell Lymphoma (MAVORIC): An International, Open-Label, Randomised, Controlled Phase 3 Trial. Lancet. Oncol. 2018, 19, 1192–1204.
- Herrera, A.; Cheng, A.; Mimitou, E.P.; Seffens, A.; George, D.; Bar-Natan, M.; Heguy, A.; Ruggles, K.V.; Scher, J.U.; Hymes, K.; et al. Multimodal Single-Cell Analysis of Cutaneous T-Cell Lymphoma Reveals Distinct Subclonal Tissue-Dependent Signatures. Blood 2021, 138, 1456–1464.
- Cristofoletti, C.; Bresin, A.; Picozza, M.; Picchio, M.C.M.C.; Monzo, F.; Helmer Citterich, M.; Passarelli, F.; Frezzolini, A.; Scala, E.; Monopoli, A.; et al. Blood and Skin-Derived Sezary Cells: Differences in Proliferation-Index, Activation of PI3K/AKT/MTORC1 Pathway and Its Prognostic Relevance. Leukemia 2019, 33, 1231–1242.
- Preston, G.C.; Feijoo-Carnero, C.; Schurch, N.; Cowling, V.H.; Cantrell, D.A. The Impact of KLF2 Modulation on the Transcriptional Program and Function of CD8 T Cells. PLoS ONE 2013, 8, e77537.
- Willinger, T.; Freeman, T.; Herbert, M.; Hasegawa, H.; McMichael, A.J.; Callan, M.F.C. Human Naive CD8 T Cells Down-Regulate Expression of the WNT Pathway Transcription Factors Lymphoid Enhancer Binding Factor 1 and Transcription Factor 7 (T Cell Factor-1) Following Antigen Encounter in Vitro and in Vivo. J. Immunol. 2006, 176, 1439–1446.
- Yang, S.; Liu, F.; Wang, Q.J.; Rosenberg, S.A.; Morgan, R.A. The Shedding of CD62L (L-Selectin) Regulates the Acquisition of Lytic Activity in Human Tumor Reactive T Lymphocytes. PLoS ONE 2011, 6, e22560.
- Kulpa, D.A.; Lawani, M.; Cooper, A.; Peretz, Y.; Ahlers, J.; Sékaly, R.P. PD-1 Coinhibitory Signals: The Link Between Pathogenesis and Protection. Semin. Immunol. 2013, 25, 219–227.
- Howden, A.J.M.; Hukelmann, J.L.; Brenes, A.; Spinelli, L.; Sinclair, L.V.; Lamond, A.I.; Cantrell, D.A. Quantitative Analysis of T Cell Proteomes and Environmental Sensors during T Cell Differentiation. Nat. Immunol. 2019, 20, 1542–1554.
- Choi, J.; Goh, G.; Walradt, T.; Hong, B.S.; Bunick, C.G.; Chen, K.; Bjornson, R.D.; Maman, Y.; Wang, T.; Tordoff, J.; et al. Genomic Landscape of Cutaneous T Cell Lymphoma. Nat. Genet. 2015, 47, 1011–1019.
- Park, J.; Yang, J.; Wenzel, A.T.; Ramachandran, A.; Lee, W.J.; Daniels, J.C.; Kim, J.; Martinez-Escala, E.; Amankulor, N.; Pro, B.; et al. Genomic Analysis of 220 CTCLs Identifies a Novel Recurrent Gain-of-Function Alteration in RLTPR (p.Q575E). Blood 2017, 130, 1430–1440.
- Ungewickell, A.; Bhaduri, A.; Rios, E.; Reuter, J.; Lee, C.S.; Mah, A.; Zehnder, A.; Ohgami, R.; Kulkarni, S.; Armstrong, R.; et al. Genomic Analysis of Mycosis Fungoides and Sézary Syndrome Identifies Recurrent Alterations in TNFR2. Nat. Genet. 2015, 47, 1056–1060.
- Vaqué, J.P.; Gómez-López, G.; Monsálvez, V.; Varela, I.; Martínez, N.; Pérez, C.; Domínguez, O.; Graña, O.; Rodríguez-Peralto, J.L.; Rodríguez-Pinilla, S.M.; et al. PLCG1 Mutations in Cutaneous T-Cell Lymphomas. Blood 2014, 123, 2034–2043.
- Okkenhaug, K.; Fruman, D.A. PI3Ks in Lymphocyte Signaling and Development. Curr. Top. Microbiol. Immunol. 2010, 346, 57–85.
- Laharanne, E.; Oumouhou, N.; Bonnet, F.; Carlotti, M.; Gentil, C.; Chevret, E.; Jouary, T.; Longy, M.; Vergier, B.; Beylot-Barry, M.; et al. Genome-Wide Analysis of Cutaneous T-Cell Lymphomas Identifies Three Clinically Relevant Classes. J. Investig. Dermatol. 2010, 130, 1707–1718.
- Scarisbrick, J.J.; Woolford, A.J.; Russell-Jones, R.; Whittaker, S.J. Loss of Heterozygosity on 10q and Microsatellite Instability in Advanced Stages of Primary Cutaneous T-Cell Lymphoma and Possible Association with Homozygous Deletion of PTEN. Blood 2000, 95, 2937–2942.
- Cristofoletti, C.; Picchio, M.C.; Lazzeri, C.; Tocco, V.; Pagani, E.; Bresin, A.; Mancini, B.; Passarelli, F.; Facchiano, A.; Scala, E.; et al. Comprehensive Analysis of PTEN Status in Sézary Syndrome. Blood J. Am. Soc. Hematol. 2013, 122, 3511–3520.
- Bresin, A.; Cristofoletti, C.; Caprini, E.; Cantonetti, M.; Monopoli, A.; Russo, G.; Narducci, M.G.M.G.M.G. Preclinical Evidence for Targeting PI3K/MTOR Signaling with Dual-Inhibitors as a Therapeutic Strategy against Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2020, 140, 1045–1053.e6.
- Marzec, M.; Liu, X.; Kasprzycka, M.; Witkiewicz, A.; Raghunath, P.N.; El-Salem, M.; Robertson, E.; Odum, N.; Wasik, M.A. IL-2- and IL-15-Induced Activation of the Rapamycin-Sensitive MTORC1 Pathway in Malignant CD4+ T Lymphocytes. Blood 2008, 111, 2181–2189.
- Marzec, M.; Halasa, K.; Kasprzycka, M.; Wysocka, M.; Liu, X.; Tobias, J.W.; Baldwin, D.; Zhang,, Q.; Odum, N.; Rook, A.H.; et al. Differential effects of interleukin-2 and interleukin-15 versus interleukin-21 on CD4+ cutaneous T-cell lymphoma cells. Cancer Res. 2008, 68, 1083–1091.
- Kittipongdaja, W.; Wu, X.; Garner, J.; Liu, X.; Komas, S.M.; Hwang, S.T.; Schieke, S.M. Rapamycin Suppresses Tumor Growth and Alters the Metabolic Phenotype in T-Cell Lymphoma. J. Investig. Dermatol. 2015, 135, 2301–2308.
- Murga-Zamalloa, C.; Rolland, D.C.M.; Polk, A.; Wolfe, A.; Dewar, H.; Chowdhury, P.; Onder, O.; Dewar, R.; Brown, N.A.; Bailey, N.G.; et al. Colony-Stimulating Factor 1 Receptor (CSF1R) Activates Akt/MTOR Signaling and Promotes T-Cell Lymphoma Viability. Clin. Cancer Res. 2020, 26, 690–703.
- Abreu, M.; Miranda, M.; Castro, M.; Fernandes, I.; Cabral, R.; Santos, A.H.; Fonseca, S.; Rodrigues, J.; Leander, M.; Lau, C.; et al. IL-31 and IL-8 in Cutaneous T-Cell Lymphoma: Looking for Their Role in Itch. Adv. Hematol. 2021, 2021, 5582581.
- Ferretti, E.; Corcione, A.; Pistoia, V. The IL-31/IL-31 Receptor Axis: General Features and Role in Tumor Microenvironment. J. Leukoc. Biol. 2017, 102, 711–717.
- Levidou, G.; Siakantaris, M.; Papadaki, T.; Papadavid, E.; Vassilakopoulos, T.P.; Angelopoulou, M.K.; Marinos, L.; Nikolaou, V.; Economidi, A.; Antoniou, C.; et al. A Comprehensive Immunohistochemical Approach of AKT/MTOR Pathway and p-STAT3 in Mycosis Fungoides. J. Am. Acad. Dermatol. 2013, 69, 375–384.
- Aronovich, A.; Moyal, L.; Gorovitz, B.; Amitay-Laish, I.; Naveh, H.P.; Forer, Y.; Maron, L.; Knaneh, J.; Ad-El, D.; Yaacobi, D.; et al. Cancer-Associated Fibroblasts in Mycosis Fungoides Promote Tumor Cell Migration and Drug Resistance through CXCL12/CXCR4. J. Investig. Dermatol. 2021, 141, 619–627.e2.
- Hong, C.H.; Lin, S.H.; Lee, C.H. CCL21 Induces MTOR-Dependent MALAT1 Expression, Leading to Cell Migration in Cutaneous T-Cell Lymphoma. In Vivo 2019, 33, 793–800.
- Narducci, M.G.; Scala, E.; Bresin, A.; Caprini, E.; Picchio, M.C.; Remotti, D.; Ragone, G.; Nasorri, F.; Frontani, M.; Arcelli, D.; et al. Skin Homing of Sézary Cells Involves SDF-1-CXCR4 Signaling and down-Regulation of CD26/Dipeptidylpeptidase IV. Blood 2006, 107, 1108–1115.
- Picchio, M.C.; Scala, E.; Pomponi, D.; Caprini, E.; Frontani, M.; Angelucci, I.; Mangoni, A.; Lazzeri, C.; Perez, M.; Remotti, D.; et al. CXCL13 Is Highly Produced by Sézary Cells and Enhances Their Migratory Ability via a Synergistic Mechanism Involving CCL19 and CCL21 Chemokines. Cancer Res. 2008, 68, 7137–7146.
More
