Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Lindsay Dong and Version 1 by Alexandre Vetcher.
Nucleic Acid (NA) aptamers are oligonucleotides. They are unique due to their primary (nucleotide sequence) and, therefore, secondary and tertiary structure. The structure of the previous order defines the propensity to form the successive one. In other words, primary (sequence) defines secondary (ability to adopt certain structures at certain conditions). The secondary define the tertiary one by means of affinity and specificity. The length of aptamers is similar to the length of PCR primers, and therefore the technology for the primers manufacturing is useful for aptamers synthesis.
- oligonucleotides
- nucleic acids
- nanotubes
- nanobiosensors
- aptamer aptasensor(s)
1. Fractionation of Nanoobjects
siRNA-Based Diagnostics
1.1. siRNA-Based Diagnostics
siRNA is a 21–25 nucleotide long RNA duplex with two unpaired overhanging nucleotides at the 3′ ends. Each chain has a phosphate group at the 5′ end and a hydroxyl group at the 3′ end.
There is a new perspective of delivering siRNA into wrapped DNA/RNA-containing nanoparticles [7][1] called “DNA baskets”. In binary DNA/RNA nanoparticles, DNA will serve as a biodegradable “basket”, which is needed to enhance its interaction with the miRNA duplex of a certain sequence. The “DNA basket” will contribute to the impossibility of miRNA degradation in the bloodstream and will serve as the basis for the placement of covalently linked ligands (i.e., targeting protein fragments or antibodies) that direct the DNA basket with its miRNA load to certain types of human body, cells. DNA-miRNA nanoparticles can be composed as dsRNA-DNA triplexes. This allows the use of additional covalently bound components that modify the structural features of the DNA basket and promote the biocompatibility of the therapeutic nanoparticle.
Non-Viral Nanoscale Delivery of Antisense Oligonucleotides
1.2. Non-Viral Nanoscale Delivery of Antisense Oligonucleotides
Such therapy does not allow the development of resistance of cancer cells and increases the effectiveness of chemotherapy, which cannot be achieved with the separate use of individual components. This represents the basis for a new type of cancer therapy based on the simultaneous delivery of an anti-cancer drug and suppressor of hypoxia-inducible factor 1α (HIF1A) [8][2]. The good impact of cancer chemotherapy is limited by the development of a large tumor drug resistance. An important mechanism of cancer resistance to chemotherapy is the adaptation of cancer cells to tumor hypoxia. Hypoxia occurs in large tumors from the early stages of tumor development as a result of insufficient oxygen supply due to exponential cell proliferation and blood supply. As a result, glucose deprivation and oxygen deficiency play a bimodal role in continued tumor development and survival. In large cases, hypoxia is an unfavorable prognostic sign of cancer, as it is associated with tumor progression and resistance to therapy.2. Different Types of Nanobiosensors
2.1. Aptamer Folding
Some types of RNA enzymes (ribozymes) have been found in biological systems and bred in the laboratory. The variety of RNA enzymes and the similarities between DNA and RNA mean that DNA can also function as an enzyme. However, this DNA enzyme has not been found in nature. The decision was made to adopt the metal-dependent DNA enzyme using the in vitro selection methodology shown in Figure 21.
Figure 21.
Sensor based on aptamer’s folding.
2.2. DNA Aptamers for Therapy and Diagnostics
Since the first monoclonal antibody was obtained in the 1970s, antibodies have been successfully and widely used in the diagnosis and therapy of human diseases. Aptamers are expected to achieve the same success as antibodies. Due to their stability, low cost and easy manipulation, DNA aptamers will continue to be actively studied and applied. RNA aptamers possess increased stability. RNA aptamers have the superiority in providing heterogeneous 3D structures, which is useful for selecting aptamers with high affinity for difficult targets needed for disease therapy. DNA aptamers may have more promising applications in in vivo diagnostics, while RNA aptamers may have promising applications in therapy. It is believed that in medicine, therapy and diagnostics based on DNA aptamers have great potential for wide application. Before they can be widely applied, therefore, there are still many problems that require further analysis in the future [12][3].2.3. Impedimetric Biosensors
The flexible array lactate biosensor in Figure 32 is made on the immobilization of l-lactate dehydrogenase (LDH) and nicotinamide adenine dinucleotide (NAD+) on a nickel oxide (NiO) film, the average sensitivity of which is increased by using graphene oxide (GO) and a magnetic field (MB beads). With GO and MB, it shows very good sensitivity (45.397 mV/mM) with a linearity of 0.992 from 0.2 mM to 3 mM. According to the results of electrochemical impedance spectroscopy (EIS), the electron transfer resistance of the LDH-NAD+-MBs/GPTS/GO/NiO film was much lower than that of the LDH-NAD+/GPTS/GO/NiO and LDH-NAD+/GPTS films/NiO, and it showed good electron transfer ability. Research is ongoing on detection limits, anti-interference effects and bending tests.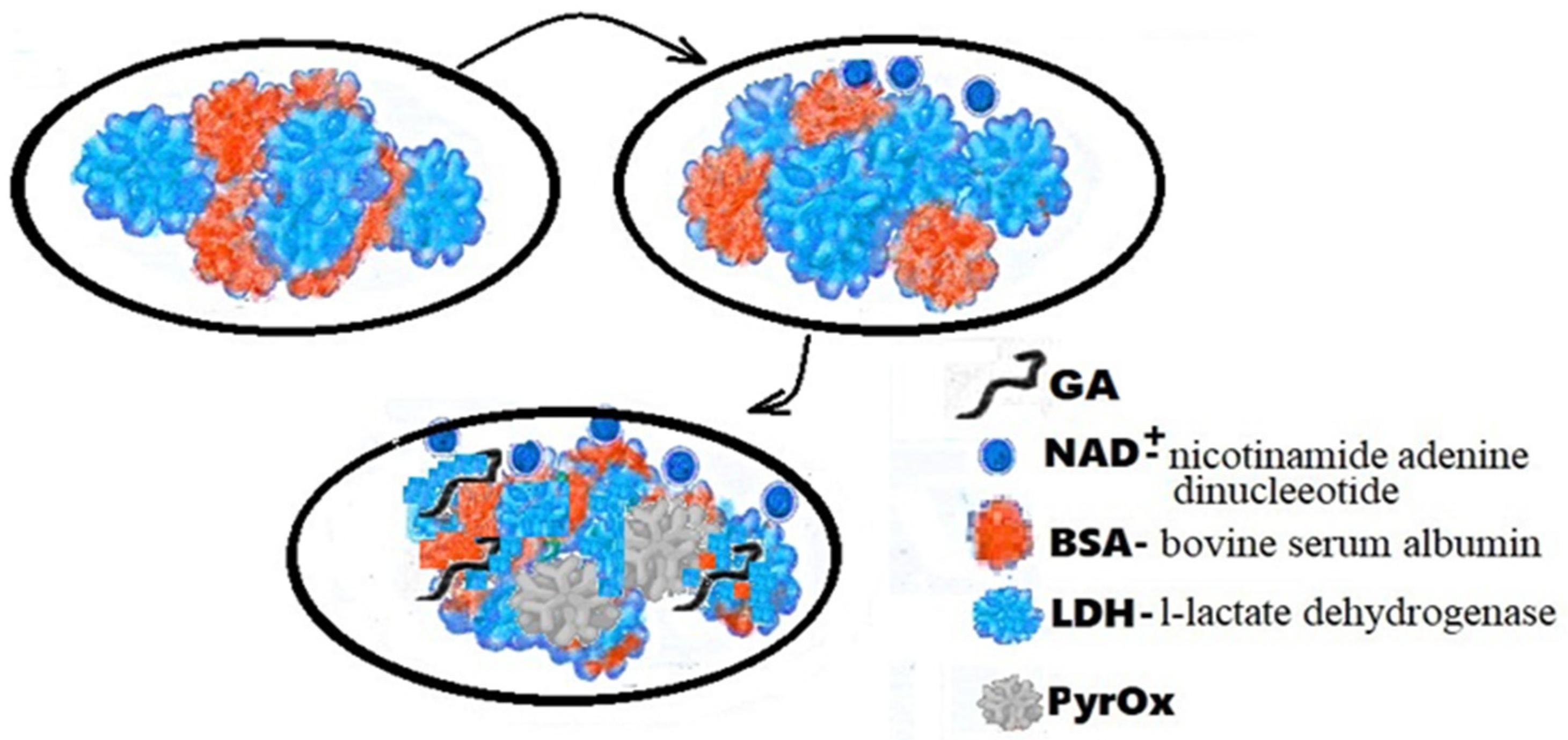
Figure 32.
Schematic representation of LDH-NAD+/PyrOx biosensor GA-BSA, LDH.
The introduction of bioreceptors in biosensors contains a limiting limit, problem immunization with small antigens and low chemical and thermal resistance. This means that there is a desire to replace bioreceptors’ artificial sensors. Molecular imprinted polymers (MMPs) are stable and reliable, in fact, which makes them easy to use at extreme pressures, temperatures, pH or organic solvents. They are still cheap to manufacture and can be kept dry. MMPs are made not only for small organic molecules: pesticides, amino acids, steroids and sugars, but also for proteins and cells.
2.4. G-quadruplex Forming Aptamer
The discovery relates to the field of nanobiotechnology and molecular medicine and concerns DNA aptamers to the thrombin exosite I interacting with prothrombin. These compounds can be used to create medicines that stop intravascular thrombus formation. Here are aptamer-based biosensors for detecting lateral flow [15][4]. Aptamers are very useful and have biorecognizing substances for the development of biosensors. Aptamers are small, single-stranded pieces of DNA used to select protein targets. A selection method (SELEX) was developed, which allows the isolation of motivated nucleic acid molecules from a large set (more than 1015) of individual molecules, called a combinatorial library [16,17][5][6]. Some aptamers in Figure 53 have every chance of not only recognizing their own targets, but also to destroy their biological vigor.
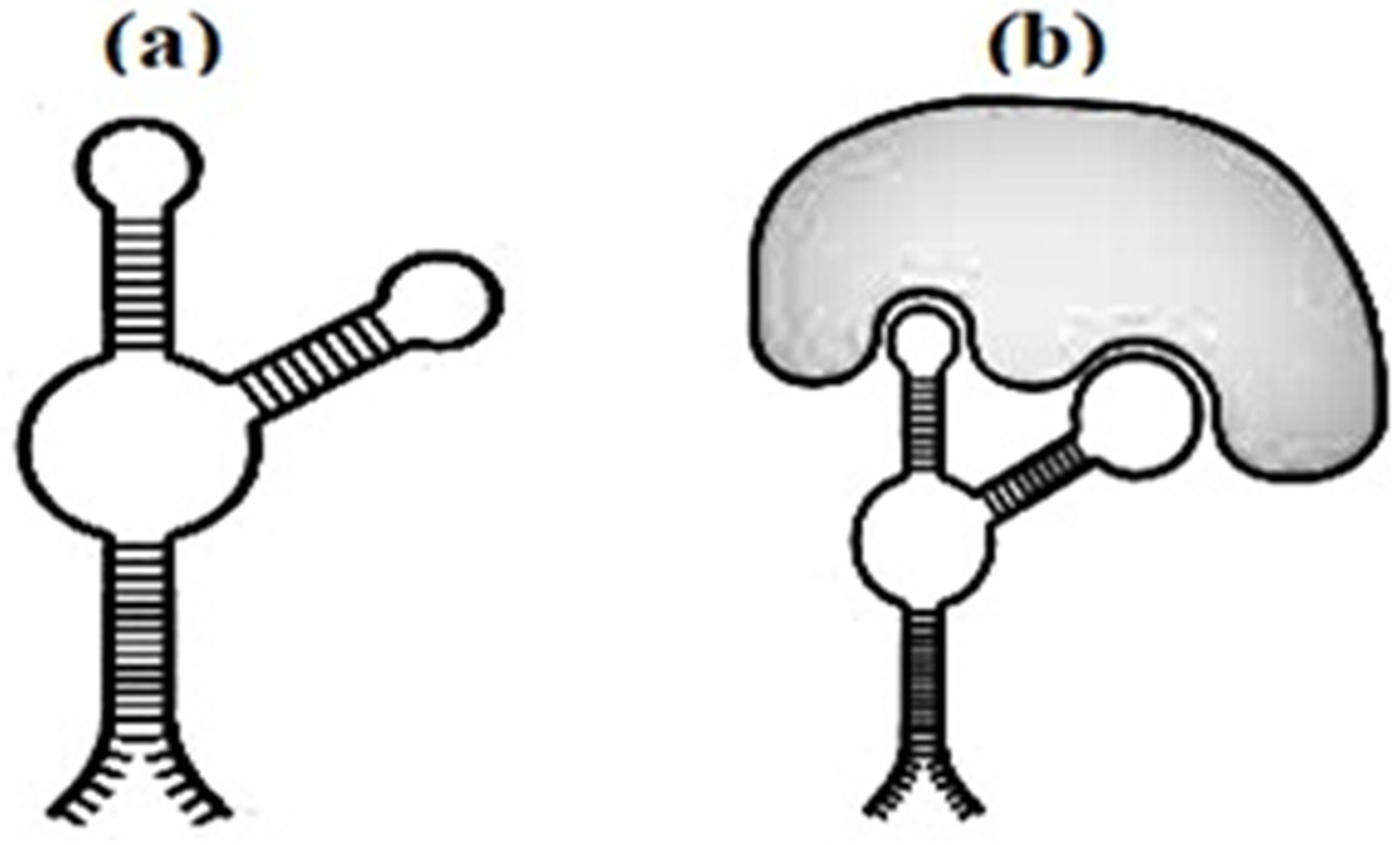

Figure 53.
The secondary structure of the aptamer (
a
) and aptamer with marker (
b
).
2.5. High-Performance Screening of SELEX RNA Aptamer with FACS Using Liposomes
The SELEX method is a method to identify and select the aptamers. It occurs by exponential enrichment in the systematic evolution of ligands. The library of oligonucleotides is gradually enriched with sequences with increased affinity for the target molecule. In classical SELEX [21][7], the process of creating target incubation, elution and amplification of the binding sequence is repeated until the vast majority of the stored pool consists of target binding sequences. Screening of the RNA aptamer by SELEX with FACS takes place using liposomes. For FACS-SELEX [22][8], one or more fluorescently labeled antibodies with specificities for desired subpopulations can be used for the enriched separation of the aptamer-bound fraction by FACS, allowing for increased specificity of selection. Since aptamers bind non-specifically to dead cells, cell-SELEX is susceptible to triggering non-specific bindings of aptamers to dead cells, and skewing or biasing the selection process. However, the combination of SELEX pre-enrichment by aptamer screening using FACS [23][9] allows you to select aptamers with the ability to light up.
2.6. Ultrasensitive and Highly Specific Recognition Aptasensors with Different Detection
Such aptasensors set the precedent that single-stranded nucleic acids quickly and firmly bind to graphene sheets using non-covalent π-π stacking and hydrophobic interactions between aromatic nitrogen-containing causes in aptamers and sp-bonded carbon atoms in graphene. In this mode of non-covalent binding, aptamers have every chance to stay on the plane of the graphene sheet, as a result of which any dye conjugated to the aptamer is located in a specific proximity to graphene. The fluorescent dye label is quenched by graphene supported by a nonradiative dipole–dipole bond. In associated targets, aptamers bind with the highest affinity and fold into specific structures. The aptamer-target complex [24][10] would disrupt the interaction between the aptamer and graphene, resulting in their release from the graphene sheet. This will increase the distance between the donor (dye) and quencher (graphene), leading to the restoration of the fluorescent signal. Switching fluorescent signals from the off state before the target changes to the on position after detecting a motivated aptamer provides a “signal” to the aptasensors for analyte detection. In good FRET [25,26,27][11][12][13] graphene aptasensors, fluorescence recovery is based on a 1:1 binding strategy since one target molecule triggers the release of one labeled aptamer from graphene. The detected fluorescence will become proportional to the concentration of motivated molecules. Subpopulations of RNA [28][14] molecules for more detailed information that specifically bind to various organic dyes are selected from a population of randomly sequenced RNA molecules [29][15].Aptamers showed some original properties, in the amount of long shelf life, simple transformation to ensure covalent bonds with material surfaces, minor batch configurations, profitability and low susceptibility to denaturation. These features have led to the necessary efforts to develop detectors based on aptamers in Figure 64, popular as aptasensors [30][16], which are designated as optical, electronic and mass-sensitive depending on the signal transmission mode.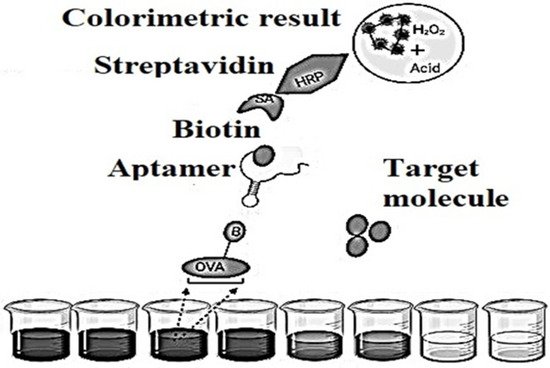


Figure 64.
Schematic representation of an indirect competitive enzyme-linked aptamer assay (ELAA)-based aptasensor.
2.7. DNA Origami as a Nanosensor
Folding of DNA molecules DNA origami is needed in a nanosensor to analyze the enzymatic energy of hAGT DNA repair. The method uses conformational configurations that cause the interaction of α-thrombin with DNA APTAMERS, and illustrates the introduction of DNA origami in Figure 75 as a biosensor for protein repair [34][17].
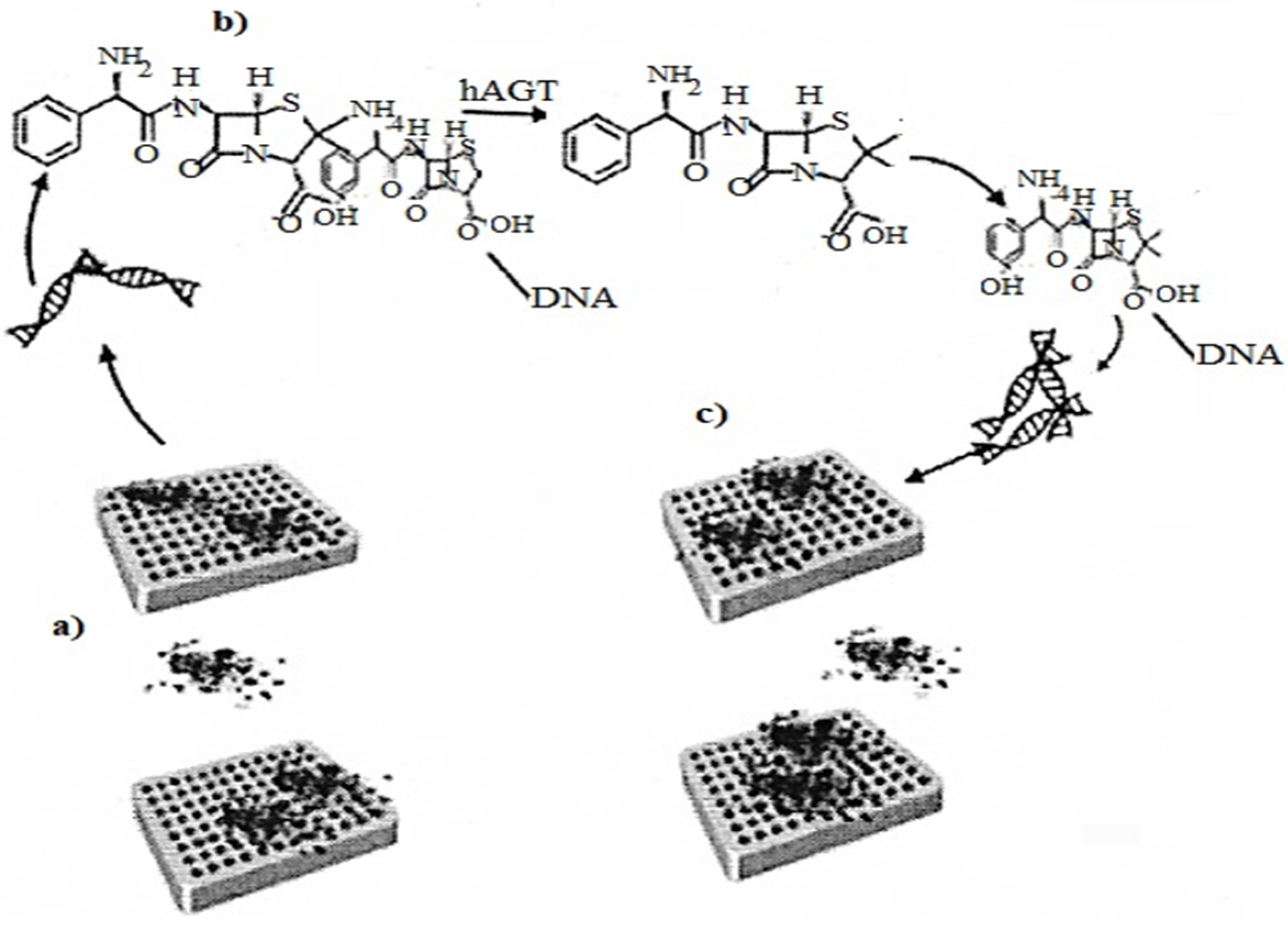

Figure 7. (a) Representation of the asymmetric binding of a-thrombin to TBA aptamers of methylated DNA origami; (b) Methyl-TBA repair by hAGT, thus G-quadruplex formation; (c) Representation of the symmetric binding of a α-thrombin to the repaired DNA origami quadruplexes.
2.8. G-quadruplex Thrombine-Binding DNA Aptamers
31-mer aptamer of DNA oligonucleotide RA36 more effectively inhibits the coagulant activity of thrombin compared to the well-known aptamer 15TGT (thrombin-binding aptamer). The RA36 aptamer has a two-pattern structure that includes two G-rich regions capable of forming a G-quadruplex. The circular dichroism method shows that the RA36 aptamer forms an antiparallel G-quadruplex similar to the 15TGT G-quadruplex. The thermal stability of the G-quadruplex RA36 is significantly lower than that of 15TGT under physiological conditions (the concentration of the stabilizing cation is 5 mM). The double quadruplex structure of RA36 is confirmed by the CD spectra of the deletion mutants, i.e., the G-quadruplex can be formed by both the first and the second G-rich site of the RA36 aptamer [36][18].
2.9. G-quadruplex Antithrombin Aptamers
In the presented objects, wescholars used the simplest 15-mer G-quadruplex antithrombin aptamers [37,38][19][20] HD1 and its analogs, namely RA36 as a covalent dimer of HD1 and GL2-HD1 as a non-covalent dimer with a G-quadruplex lock. HD1 and RA36 are single-stranded G-quadruplexes, while GL2-HD1 forms non-covalent (GL2-HD1)2 dimers in a concentration dependent manner, giving ~50% dimers at 1 mmol dm−3. The oligomerization of aptamer HD1 affects its pharmacokinetics, and therefore, requires a thorough consideration during the development of new aptamer-based antithrombotic drugs. In general, a simple and robust technique is necessary for the quantification of various oligomeric and conformational states of G-quadruplexes. The currently used approach is the size exclusion high performance liquid chromatography (SE HPLC); however, its common protocols lack a linear correlation between the chromatographic retention volume and the logarithm of molecular weight for monomeric G-quadruplexes, the reason being probably the sub-optimal separation conditions. As a result of such complications, the molecular mass of aptamer HD1 was twice overestimated. Analytical ultracentrifugation is used as an additional method for the estimation of G-quadruplexes oligomeric [39][21].2.10. NA Aptamers with GFP Chromophore for Biosensors
In cell and molecular biology, the GFP gene is often used as an expression reporter. It has been used in modified forms to create biosensors, and many animals have been created that express GFP, demonstrating that the gene can be expressed throughout a given organism, in particular organs or cells of interest. GFP can be introduced into animals or other species by transgenic methods and stored in their genome and the genome of their offspring. To date, GFP is expressed in many species, including bacteria, yeast, fungi, fish and mammals, including human cells. Biosensors based on GFP fusion proteins are powerful tools for real-time observation of events in living cells. Insertion of GFP into another protein has made it possible to obtain biosensors capable of signaling intracellular events through intrinsic fluorescence changes, resonant fluorescence energy transfer and changes in subcellular localization. The difficult task of finding the correct insert site to obtain a biosensor can be accelerated by screening libraries of random GFP inserts [40][22].2.11. DNA Aptamers Binding to M11
The in vitro method directly uses live bacterial cells and the strategy of systematic ligand evolution by exponential enrichment (SELEX). Individual aptamer sequences, if screened based on their binding to 10 M-types, can be used as targets. Aptamer pools obtained during 5–8 rounds of SELEX showed high affinity for S. pyogenes target cells. Several aptamer sequences preferentially bind to the M11 M-type of S. pyogenes. The improved SELEX method with bacterial cells has been successfully used to generate aptamers that are selective for S. pyogenes and some of its M-types. These aptamers are potentially useful for detecting S. pyogenes, deriving binding profiles of various M-types, and developing new M-typing technologies for non-specialist laboratories or point-of-care testing [41][23].2.12. Impedance Detection Methods for Miniature Analytical Systems
Sensor systems based on impedance detection can be divided into two types depending on the presence or absence of a biorecognition element in them. Systems of the first type, which are called impedimetric biosensors (ID-biosensors), fix the change in impedance caused by the binding of the target and bioligand (antibody, antimicrobial peptide or aptamer) on the surface of the electrodes. Detectors of the second type are called impedimetric sensors (ID-sensors), and register the change in electrical impedance as a result of chemical and metabolic processes occurring in the sample volume and directly on the surface of the electrodes. System impedance analysis detection as efficient and effective method in miniature analytical systems has a number of developments in the field of sensors based on impedance detection and the possibility of integrating them in laboratories on a chip (LNC) [42][24].References
- Minko, T.; Khandare, J.J.; Vetcher, A.A.; Soldatenkov, V.A.; Garbuzenko, O.B.; Saad, M.; Pozharov, V.P. Multifunctional nan-otherapeutics for cancer. In Multifunctional Pharmaceutical Nanocarriers; Part of Fundamental Biomedical Technologies; Springer: New York, NY, USA, 2008; Volume 4, pp. 309–336.
- Wang, Y.; Saad, M.; Pakunlu, R.I.; Khandare, J.J.; Garbuzenko, O.B.; Vetcher, A.A.; Soldatenkov, V.A.; Pozharov, V.P.; Minko, T. Nonviral nanoscale-based delivery of antisense oligonucleotides targeted to hypoxia-inducible factor 1α enhances the efficacy of chemotherapy in drug-resistant tumor. Clin. Cancer Res. 2008, 14, 3607–3616.
- Guan, J.G.; Miao, Y.Q.; Zhang, Q.J. Impedimetric biosensors. J. Biosci. Bioeng. 2004, 97, 219–226.
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479.
- Huang, L.; Tian, S.; Zhao, W.; Liu, K..; Ma, X.; Guo, J. Aptamer-based lateral flow assay on-site biosensors. Biosens. Bioelectron. 2021, 186, 113–279.
- Ayela, C.; Roquet, F.; Valera, L.; Granier, C.; Nicu, L.; Pugnière, M. Antibody-antigenic peptide interactions monitored by SPR and QCM-D. A model for SPR detection of IA-2 autoantibodies in human serum. Biosens. Bioelectron. 2007, 22, 3113–3119.
- Wu, Y.; Midinov, B.; White, R.J. Electrochemical sensor based on aptamer for real-time monitoring of insulin. ACS Sens. 2019, 4, 498–503.
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Liu, B.; Lu, A. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358.
- Mayer, G.; Ahmed, M.-S.L.; Dolf, A.; Endl, E.; Knolle, P.A.; Famulok, M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010, 5, 1993–2004.
- Dejeu, J.; Van der Heyden, A.; Spinelli, N.; Defrancq, E.; Coche-Guérente, L. Recent progress in the design of G-quadruplex–based electrochemical aptasensors. Curr. Opin. Electrochem. 2021, 30, 100812.
- Huang, R.R.; He, V.; Xia, Y.Y.; Xu, X.P.; Liu, K.; Xie, H.; Wang, S.; Peng, L.D.; Liu, Y.F.; Liu, Y.; et al. Sensitive aptasensor strategy-based signal amplification using hemin. Small 2019, 15, 1–7.
- Deiminat, B.; Runagi, G.H. A novel visible light photoelectrochemical aptasensor for determination of bisphenol A based on surface plasmon resonance of gold nanoparticles activated g-C3N4 nanosheets. J. Electroanal. Chem. 2021, 886, 115–122.
- Hosseinzadeh, L.; Mazloum-Ardakani, M. Chapter Six—Advances in Aptasensor Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 99, pp. 237–279.
- Zhang, H.; Han, Y.; Guo, Y.; Dong, C. Porphyrin functionalized graphene nanosheets-based electrochemical aptasensor for la-bel-free ATP detection. J. Mat. Chem. 2012, 22, 23900–23905.
- Zhao, J.; Zhai, Q. Recent advances in the development of ligands specifically targeting telomeric multimeric G-quadruplexes. Bioorg. Chem. 2020, 103, 104229.
- Nguyen, T.Q.N.; Lim, K.W.; Phan, A.T. Folding Kinetics of G-Quadruplexes: Duplex Stem Loops Drive and Accelerate G-Quadruplex Folding. J. Phys. Chem. B 2020, 124, 5122–5130.
- Sun, D.P.; Lin, X.A.; Lu, J.; Wei, P.; Luo, Z.B.; Lu, X.E.; Chen, Z.G.; Zhang, L.Y. DNA nanotetrahedron-assisted electro-chemical aptazensor for the detection of cardiac troponin I based on the joint catalysis of hybrid nanoenzyme, natural enzyme and artificial DNAzyme. Biosens. Bioelectron. 2019, 142, 15.
- Yuminova, A.V.; Smirnova, I.G.; Arutyunyan, A.M.; Kopylov, A.M.; Golovin, A.V.; Pavlova, G.V. The structure of G-quadruplex thrombine-binding DNA aptamer RA36. Mosc. Univ. Chem. Bull. 2015, 70, 43–46.
- Alieva, R.R.; Zavyalova, E.G.; Tashlitsky, V.N.; Kopylov, A.M. Quantitative characterization of oligomeric state of G-quadruplex antithrombin aptamers by size exclusion HPLC. Mendeleev Commun. 2019, 29, 424–425.
- Kretz, C.K.; Cuddy, K.R.; Stafford, A.R. HD1, a thrombin- and prothrombin-binding DNA aptamer, inhibits thrombin gener-ation by attenuating prothrombin activation. Thromb. Haemost. 2010, 103, 83–93.
- Close, D.M.; Ripp, S.; Sayler, G.S. Reporter Proteins in Whole-Cell Optical Bioreporter Detection Systems, Biosensor Integrations, and Biosensing Applications. Sensors 2009, 9, 9147–9174.
- Hamula, C.L.; Peng, H.; Wang, Z.; Tyrrell, G.J.; Li, X.-F.; Le, X.C. An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes. Methods 2015, 97, 51–57.
- Yuan, X.; Hu, X.; Sun, Y.; Wu, Z.; Lu, P. A weak grid impedance detection method based on windowed Fourier transformation. Power Syst. Prot. Control 2018, 46, 96–101.
- Challier, L.; Miranda-Castro, R.; Barbe, B.; Fave, C.; Limoges, B.; Peyrin, E.; Ravelet, C.; Fiore, E.; Labbé, P.; Coche-Guérente, L.; et al. Multianalytical Study of the Binding between a Small Chiral Molecule and a DNA Aptamer: Evidence for Asymmetric Steric Effect upon 3′- versus 5′-End Sequence Modification. Anal. Chem. 2016, 88, 11963–11971.
More
