Polyploidy events have long been recognized as the primary driving force behind the survival of the vast majority of plant lineages, playing critical roles in crop evolution, speciation, and domestication. The fascinating modern genomics era has revealed that the genomes of all flowering plants have been polyploidized multiple times. To help safeguard the future of the global food supply in the face of climate change, a thorough understanding of polyploid genomes is critical, especially for the improvement of members of the grass family, Poaceae, which includes the world’s big three cereals—rice, wheat, and maize—as well as some potential underutilized ancient grasses (such as teff) that are less well known or studied.
1. Introduction
The scientific community has long sought to understand how some species were able to cross the Cretaceous–Paleogene (K–Pg, or formerly K–T) boundary, surviving the catastrophic mass extinction while adapting to an ever-changing world
[1][2][1,2]. Interestingly, recent paleontological research has revealed that the evolution of South American rainforests, from open canopies with dense stands of gymnosperms to closed canopies dominated by angiosperms, was strongly influenced by the asteroid’s blast
[3]. Based on the analysis of thousands of fossilized plant remnants collected from the hyper-diverse tropical rainforests, it was determined how plant communities, particularly angiosperms, recovered and thrived after the mass extinction, with increased soil nutrient availability being one of the main contributing factors
[3]. Most angiosperms, which account for approximately 80% of today’s flora, have duplicated their genomes at some point during their evolution. These species are termed polyploids, possessing multiple copies of the same genome
[3][4][3,4]. According to Fawcett et al.
[4], the ancient doubling of the genome in these species, which peaked between 60 and 70 million years ago, provided an advantage in adjusting to dramatic environmental change, which killed off the less well-endowed species.
2. Impacts of Polyploidy at a Glance
Polyploidy, which results from an organism’s or cell’s genome duplication, can have a significant impact on various levels of biological organization
[5][21]—from genes to entire ecosystems (
Figure 1). Polyploidy is generally divided into two types: autopolyploidy and allopolyploidy, which involve intraspecific WGD and interspecific hybridization events, respectively
[6][7][22,23]. Autopolyploids typically have lower fertility, whereas allopolyploids have the potential for hybrid vigor (or heterosis)
[8][24]. Much research effort has been directed toward allopolyploids to understand the consequences of polyploidy; however, relatively little is known about the outcomes of polyploidy in autopolyploids
[9][10][5,16]. Genetic alterations to the basic omes, such as the genome and transcriptome (
Figure 1), are almost always unavoidable in nascent polyploids, and usually only those with dynamism can survive
[10][11][16,17]. Over the last three decades, it has become clear that relatively young allopolyploids may have a highly dynamic genome, undergoing “genomic shock”, where the genome experiences a variety of phenomena such as structural changes and massive reorganization to cope with disturbances
[12][13][25,26]. Even though numerous studies on crops and their close relatives have revealed various genetic phenomena associated with polyploidy, there is a gap in understanding commonalities derived from shared polyploid cellular processes, particularly across different fields of study
[5][21]. Polyploidy, as a biological force (
Figure 1), is poised to improve crop fitness and diversity in the face of climate change as the impacts and consequences of polyploidy on adaptability and stress resilience in plants becomes better understood. The increase in an organism’s cellular ploidy caused by genome replication without mitosis, known as endopolyploidy, has been shown to play important roles in physiology and development via cellular, metabolic, and genetic effects
[14][27].
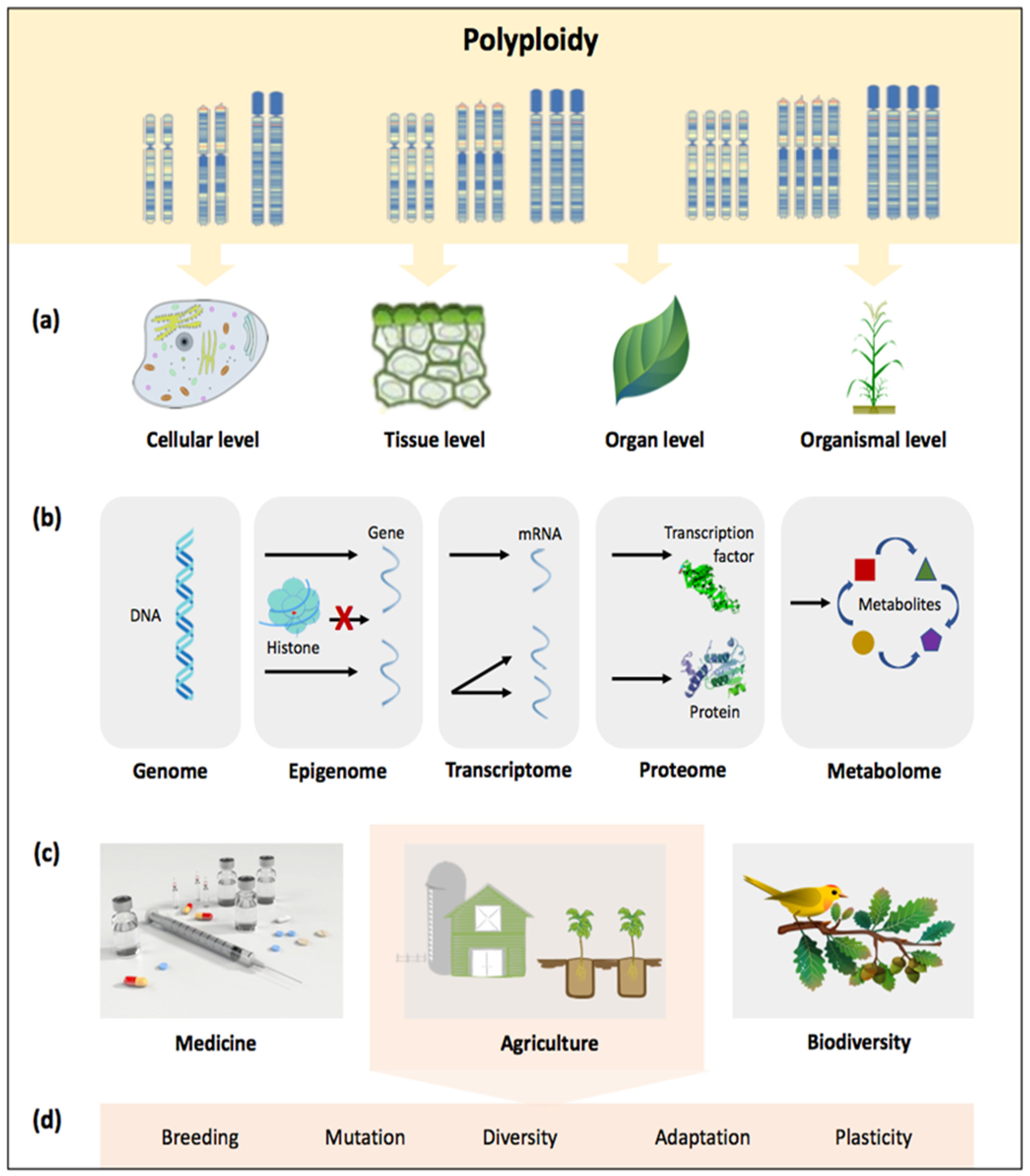
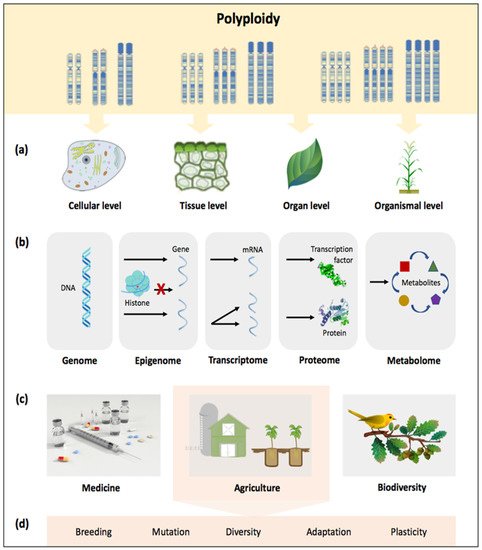
Figure 1. Advances in understanding polyploidy, with its role and implications from cells to ecosystems; (a) polyploidy involves the fusion of two or more genomes within one nucleus, affecting cells, tissues, organs, and organisms; (b) polyploidy as the product of a dynamic process in which restructuring of the genome, epigenome, transcriptome, proteome, and metabolome occurs through various changes; polyploidy as a biological force in (c) various fields and more specifically, (d) agriculture.
2.1. Diversity and Evolutionary Dynamics
The K–Pg boundary was a pivotal period in Earth’s history, wiping out half of all species and drastically altering the planet’s environment. Polyploidy, followed by gene loss and diploidization, has long been recognized as a significant evolutionary force in plants and other organisms, facilitating diversity and rapid speciation
[15][16][17][28,29,30]. For millions of years, events associated with whole-genome duplications have been a major contributor to the success of angiosperms. However, evidence for proposed ancient polyploidy events prior to the divergence of monocots and eudicots is ambiguous, especially in analyses of conserved gene order
[3]. Many plant lineages, including monocots (such as rice) and eudicots (such as Arabidopsis), have had at least one paleo-polyploidy event. The determination of phylogenetic placements and timing of polyploidy has been the most pressing issue in plant evolution, with highly contradictory lines of evidence indicating that many polyploids and their phylogenetic positions remain unknown
[17][18][30,31].
Domestication of wild plants around 10,000 years ago was thought to be the key innovation that led to human population expansion, which has since been sustained through continued selection and breeding of crops with novel desirable phenotypes. These phenotypes, which distinguish the domesticated crop from its wild ancestors, frequently form a group of improved traits known as “domestication syndrome”
[19][32]. Seed dispersal, increased seed size, loss of seed dormancy and shattering, and fragrance are the examples of “domestication syndrome”
[20][33]. The grass family, Poaceae, which includes all the major cereals, such as wheat, maize, and rice, provides a cohesive system for crop domestication studies, with the clade thought to have emerged around 75 million years ago, leading to cereal-specific lineages
[20][21][33,34]. It is important to note that a genome duplication event shared by all grass crops occurred around 70 million years ago, prior to grass divergence
[22][35]. Following that, certain cereals, such as wheat and maize, experienced additional lineage-specific polyploidy events, resulting in significant functional diversity for domestication and adaptation when selective fractionation occurred
[23][36].
While the early stage studies of polyploidy were primarily based on the synthetic autopolyploid, Arabidopsis, much understanding of crop genome evolution following polyploidy has come from research on wheat, an allopolyploid
[10][11][16,17]. Allopolyploidy was discovered to limit the chromosome number and the distribution of low-copy sequences in the hexaploid wheat genome, in comparison to those seen in its diploid progenitors (A, B, and D subgenomes), demonstrating a loss of homoeologous regions
[23][24][36,37]. Polyploidy has facilitated many domestication-related and evolutionary traits in allopolyploid wheat over the last few decades, with notable examples being the fresh-threshing character
[25][38] and nitrogen (N) uptake and assimilation
[26][27][39,40].
Crop species in the Poaceae family, other than wheat, have been successful in surviving or thriving in a variety of environmental conditions, albeit with limitations in adaptive potential due to domestication, particularly of those with large genomes. Targeted selection and demographic bottlenecks during domestication are thought to have compromised adaptive variation, potentially reducing the extent of phenotypic parallelism observed during adaptation
[20][28][33,41]. Nonetheless, there is a lack of understanding of parallel adaptation within crop species, which is critical in improving modern breeding
[20][33]. Understanding polyploidy in non-crops or invasive species, such as weeds and Spartina, is also important for improving current agricultural management practices that cause rapid environmental change
[29][30][42,43]. Even though plants in the Poaceae family are the dominant primary producers and the foundation of terrestrial ecosystems, the macroevolutionary dynamics of plant clades across the K–Pg boundary have received far less research attention than the prominent vertebrate clades
[16][17][29,30].
2.2. Crop Improvement
In general, polyploidization in plants involves two distinct processes: genome merger and genome doubling (multiplication)
[31][44]. Artificial, also known as synthetic polyploids, are frequently produced in plant breeding through a genome merger that facilitates hybridization, which involves interspecific crosses between different genotypes that typically occur within the same taxonomic species. While traditional selection is based on phenotypes, the union of different genotypes during hybridization allows interactions among differentiated alleles that result in a variety of effects (
Figure 2), the most prominent of which is heterosis, also known as hybrid vigor and outbreeding enhancement
[32][33][45,46]. Heterosis is frequently used to create a hybrid with superior characteristics such as higher yield and stress tolerance.
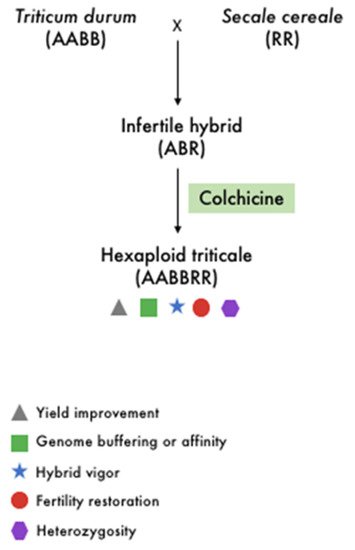
Figure 2.
Schematic representation of artificial polyploidy of wheat with some of the main implications of polyploidy in crop improvement.
The potential for epistatic effects on heterosis in allopolyploids has been widely dis-cussed and is thought to have contributed to the successful domestication of many modern allopolyploid crop species.
A recent review by Labro et al.
[34][47] provides a clear overview on heterosis and its importance in crop breeding. Given that the number of interacting gene pairs increases with ploidy level, the situation in polyploids can be complicated. Dissecting the genetic mechanisms underlying the merge is an important goal in polyploid research because this knowledge may be used to enhance phenotypic novelty and heterosis in crops
[10][33][34][16,46,47]. Nonetheless, unlike in diploids, the greater complexity of genotype and phenotype in polyploids makes it difficult for breeders to effectively link genotype to phenotype
[8][35][24,48].
For cereal crops, the first synthesized amphidiploid species is the triticale—a chromosome-doubled intergeneric hybrid obtained from the distinct cross between wheat species (
Triticum spp., AA, AABB, or AABBDD) and rye (
Secale cereale L., RR) (
Figure 2). Various genome combinations and ploidy levels have resulted from the attempted crosses, including tetraploid AARR, hexaploid AABBRR, and octoploid AABBDDRR. Triticeae synthetic allopolyploid species, which frequently involve
Triticum spp. and
Aegilops spp., are regarded as excellent models for studying polyploid evolution, particularly for the wheat–rye hybrid triticale
[36][53]. Triticale has a very complex genome compared to many other allopolyploids due to its large genome size and high ploidy level. Ploidy manipulation is a well-known and effective technique for developing novel phenotypes in cereal breeding. Nonetheless, severe seed sterility has been observed in rice since the 1930s as a barrier to autotetraploid cultivation. Recent research has identified two fertile high seed-setting autotetraploids, the Polyploid Meiosis Stability (PMeS) and Neo-Tetraploid lines, developed through tissue culture followed by chemical treatment and extensive hybridization between different autotetraploid rice followed by directional selection, respectively. Identifying the genes responsible for their high seed fertility may allow for the efficient development of autotetraploid rice varieties via marker-assisted selection (MAS) or genome editing
[37][54]. Further, research on understanding the origin of these polyploids can set the platform for successful development and deployment of polyploid rice in future.
3. Dissecting the Polyploid Potential of Current and Future Crops in the Genomic Era
Cytological analysis of chromosomes was historically used to identify polyploid plants until recent decades when advances in genomics have enabled more efficient, large-scale research on more ancient and cryptic cases of polyploidy
[38][9].
With the introduction of third generation sequencing technologies (such as long-read sequencing and scaffolding) in the late 2000s, sequencing costs were further reduced with simplified preparatory methods, allowing for longer read lengths and more comparative genome analyses across multiple species. However, these technologies have a high error rate and necessitate very high-quality DNA
[39][63]. Using the original strategy of whole genome profiling-based BAC sequencing, the first monoploid genome of a sugarcane hybrid was developed
[40][64]. The Single Molecule Real-Time (SMRT) sequencing is one of the most widely used long-read sequencing technologies, having been used to assemble various crop genomes such as quinoa (
Chenopodium quinoa) with a read length of about 12 Kbp
[41][65]. Long-range scaffolding technologies can be used for contig extension or scaffolding to improve the contiguity of an assembly. Utilization of SMRT and Hi-C sequencing technologies in combination of Illumina short-read sequencing led to the development of an allele-defined genome assembly of
Sachharum spontaneum, the wild autopolyploid progenitor of sugarcane
[42][66]. Continuous advancements in polyploid crop genome sequencing and assembly will benefit modern plant breeding and aid researchers in studying the genotype–phenotype–environment relationship. Multiple high-quality reference genomes or a pan-genome, in combination with reference sub-genomes, are critical for identifying genomic variants associated with agronomic traits of interest and for better understanding of the genomes of important species, including both major and minor crops that play a role in sustaining global food security in the face of climate change.
Advances in crop genetics and genomics have paved the way for
theour understanding of polyploidy in crops. Recent research suggests that polyploidization is enhanced in many crops, resulting in profound changes in their plant growth and cell wall composition
[25][43][38,67]. The development of a comprehensive phylogenetic framework of numerous plant genera, including many major crops, revealed that more polyploidization events occurred in domesticated crops than in wild counterparts, allowing for the formation and selection of important traits through the expansion of genetic materials
[44][68].
4. Reappraising Polyploidy for Crop Improvement in the 21st Century: The Road Ahead
Polyploidy has played a key role in crop evolution and domestication for millions of years, increasing allelic diversity and fixing heterozygosity while generating new phenotypic variations as a result of duplicated genes that give rise to new functions
[45][74]. There is no doubt that polyploidy has played a significant historical role in crop improvement, but moving forward, with the need for accelerated breeding strategies, there are significant gaps in the existing knowledge base that must be addressed, particularly in how polyploidy impacts biological processes and affects the genetic transmission of important agronomic traits between different crop species and their wild counterparts. Because polyploidy has been highly compartmentalized, the similarities between polyploids in terms of cellular processes at various levels of biological organization and diversity remain largely unexplored
[5][21]. Although it is well known that polyploidy almost always increases cell size, resulting in a decreased ratio of cell surface/volume
[46][75], the fundamental impact of these polyploidy-driven cell changes, which are frequently influenced by changing environments, is still unclear
[5][47][21,76]. This uncertainty, which stems primarily from the lack of translatable communications across disciplinary boundaries, such as genomics, ecology, evolution, and agriculture, is regarded as one of the most significant challenges in developing strategies to combat climate and/or social crises, such as sustaining biodiversity and ecosystem services and improving agricultural yield in a rapidly changing world (
Figure 3)
[5][48][13,21]. Therefore, interdisciplinary research on polyploidy involving multiple disciplines may facilitate discovery of novel insights into the million-year-old phenomenon at various levels of organization, from cells to organisms (
Figure 1).
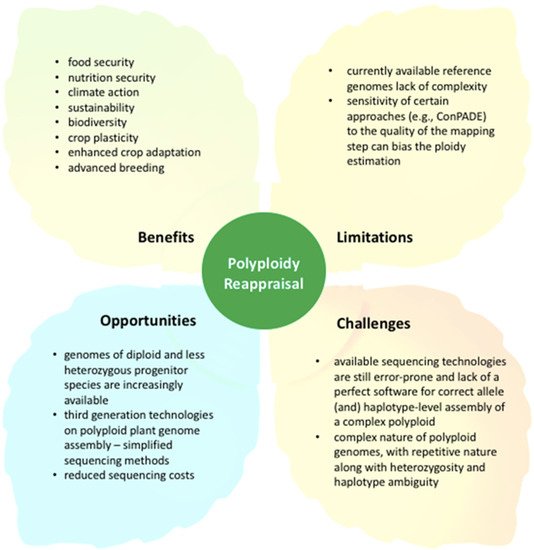
Figure 3.
Benefits, opportunities, limitations, and challenges of reappraisal of polyploidy events in crops.