Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Juli Jing.
Meiosis is an essential cell-division process for ensuring genetic diversity across generations. Meiotic recombination ensures the accuracy of genetic interchange between homolous chromosomes and segregation of parental alleles. Programmed DNA double-strand breaks (DSBs), catalyzed by the evolutionarily conserved topoisomerase VIA (a subunit of the archaeal type II DNA topoisomerase)-like enzyme Spo11 and several other factors, is a distinctive feature of meiotic recombination initiation.
- meiosis
- double-strand break (DSB)
- homologous recombination
- chromosome
- hotspots
1. Introduction
In flowering plants, reproductive cells develop in the ‘sporophytic generation’ and then differentiate into the gamete-forming ‘gametophytic generation’ [1,2][1][2]. During the reproductive process, the production of gametes requires that the genetic complement be reduced by one-half [3,4,5][3][4][5]. This specialized nuclear division, called meiosis, includes one round of DNA replication followed by two successive rounds of cell division, thereby ensuring the ploidy of the zygotic genome [6,7][6][7]. In the first meiotic division (meiosis I), homologous chromosomes are brought in close to pair and undergo synapsis, promoting the reciprocal exchange of parental chromosome fragments and thereby increasing the genetic diversity of the progeny [7,8,9][7][8][9]. Meiotic recombination is initiated by the formation of programmed DNA double-strand breaks (DSBs) catalyzed by the evolutionarily conserved type II topoisomerase–like enzyme SPO11 and several accessary proteins [9,10,11,12,13][9][10][11][12][13]. After resection, two SPO11 molecules remain covalently bound to each 5′ end of the nicked DNA, which is then processed by the MRX complex (Mre11-Rad50-Xrs2) with the cooperation of Sae2 to release the SPO11-bound nicked DNA oligonucleotide [14,15,16,17,18,19][14][15][16][17][18][19]. Following the action of the 5′ to 3′ exonuclease Exo1, the DNA ends are further degraded to produce 3′ single-stranded DNA tails [9,20,21,22][9][20][21][22].
2. Defining Meiotic DSB Hotspots in Different Species
Meiotic DSBs are not randomly distributed along eukaryotic chromosomes; rather, they are concentrated within discrete regions described as DSB hotspots [92][23]. To generate a high-resolution physical map of the meiotic DSB landscape, several methodologies have been established over the past three decades. SPO11-oligo mapping is achieved by immunoprecipitating tagged-Spo11 bound with oligonucleotides, which subsequently go through end-labeling, purification and sequencing [98][24]. This method has been effectively applied in several yeast species, mouse and Arabidopsis (Table 21) [92,99,100,101,102][23][25][26][27][28]. SSDS takes advantage of an antibody specifically recognizing either one of two DNA recombinases, RAD51 or DMC1, and utilizes chromatin immunoprecipitation to enrich single-stranded DNA that has undergone single-end invasion. With the high-throughput sequencing of enriched single-stranded DNA, the genome-wide distribution of DSB hotpots was successfully obtained in maize, mouse and human [103,104,105][29][30][31] (Table 2).Table 21.
Meiotic DSB hotspots identified in different species by SPO11-oligo mapping or single-stranded DNA sequencing (SSDS).
| Species | Genome Size | Chromosome No. | Number of DSBs | DSB Hotspot No. | Most Common DSB Location |
Average Width (kb) | Predominantly DSB Formation Among Transposon |
Method | Hotspot Detection | References |
|---|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae (SK1) | 12.1 Mb | 16 | ~175 | 3604–4099 | Gene promoters | 0.248–0.264 | Ty retrotransposons | SPO11-oligos | Enrichment threshold | [92,101,110,111][23][27][32][33] |
| S. cerevisiae (YPS128) | 12.1 Mb | 16 | ~175 | 4177 | Gene promoters | 0.265 | n/a | SPO11-oligos | Enrichment threshold | [112][34] |
| S. cerevisiae (UWOPS03-461.4) | 12.1 Mb | 16 | ~175 | 3881 | Gene promoters | 0.256 | n/a | SPO11-oligos | Enrichment threshold | [112][34] |
| S. pombe | 13.8 Mb | 3 | ~60 | 603 | All chromosome regions | 1.4 | n/a | Rec12-oligos | Enrichment threshold | [113][35] |
| M. musculus (9R×13R) | 2.8 Gb | 20 | ~250 | 9874– 15,677 |
Intergenic | ~2.000–3.400 | LTR retrotransposons SINE |
SSDS | Peak calling | [103,104][29][30] |
| M. musculus (9R) | 2.8 Gb | 20 | ~250 | 14,869 | Intergenic | ~2.000 | n/a | SSDS | Peak calling | [104][30] |
| M. musculus (13R) | 2.8 Gb | 20 | ~250 | 15,481 | Intergenic | ~2.000 | n/a | SSDS | Peak calling | [104][30] |
| M. musculus (B6) |
2.8 Gb | 20 | ~250 | 18,313 | Intergenic | ~2.000 | n/a | SSDS | Peak calling | [104][30] |
| M. musculus (B6) |
2.8 Gb | 20 | ~250 | 13,960 | Intergenic | ~0.281 | n/a | SPO11-oligos | Enrichment threshold | [100][26] |
| Arabidopsis thaliana | 135 Mb | 5 | ~250–300 | 5914 | Gene promoters and terminators | 0.823 | Helitron /Pogo/Tc1/Mariner DNA transposons | SPO11-1-oligos | Peak calling | [99][25] |
| Zea mays | 2.4 Gb | 10 | ~500 | 3126 | All chromosome regions | 1.2 | Gypsy retrotransposons | SSDS | Peak calling | [105][31] |
3. Control of Meiotic DSB Formation by Protein Phosphorylation
3.1. Cyclin-Dependent Kinases (CDKs)
CDKs in conjunction with their cyclin partners represent an ancient molecular switch that promotes and regulates cell-cycle progression [4,114,115][4][40][41]. The fundamental theme of how CDKs mediate meiotic recombination initiation was mostly drawn from studies in yeast. In S. cerevisiae, the activation of Cdc28 by its two B-type cyclin partners, Clb5 and Clb6, stimulates the phosphorylation on Mer2 during pre-meiotic DNA replication (Figure 1A) [115,116,117,118,119][41][42][43][44][45]; subsequently, phosphorylated Mer2 recruits other Spo11-accessary proteins to initiate DSB formation (Figure 1B) [45,120,121][46][47][48]. Similarly, in S. pombe, the association between Cdc2 and any of the three cyclins, Crs1, Cig1 and Cig2, is crucial for DSB formation [114][40].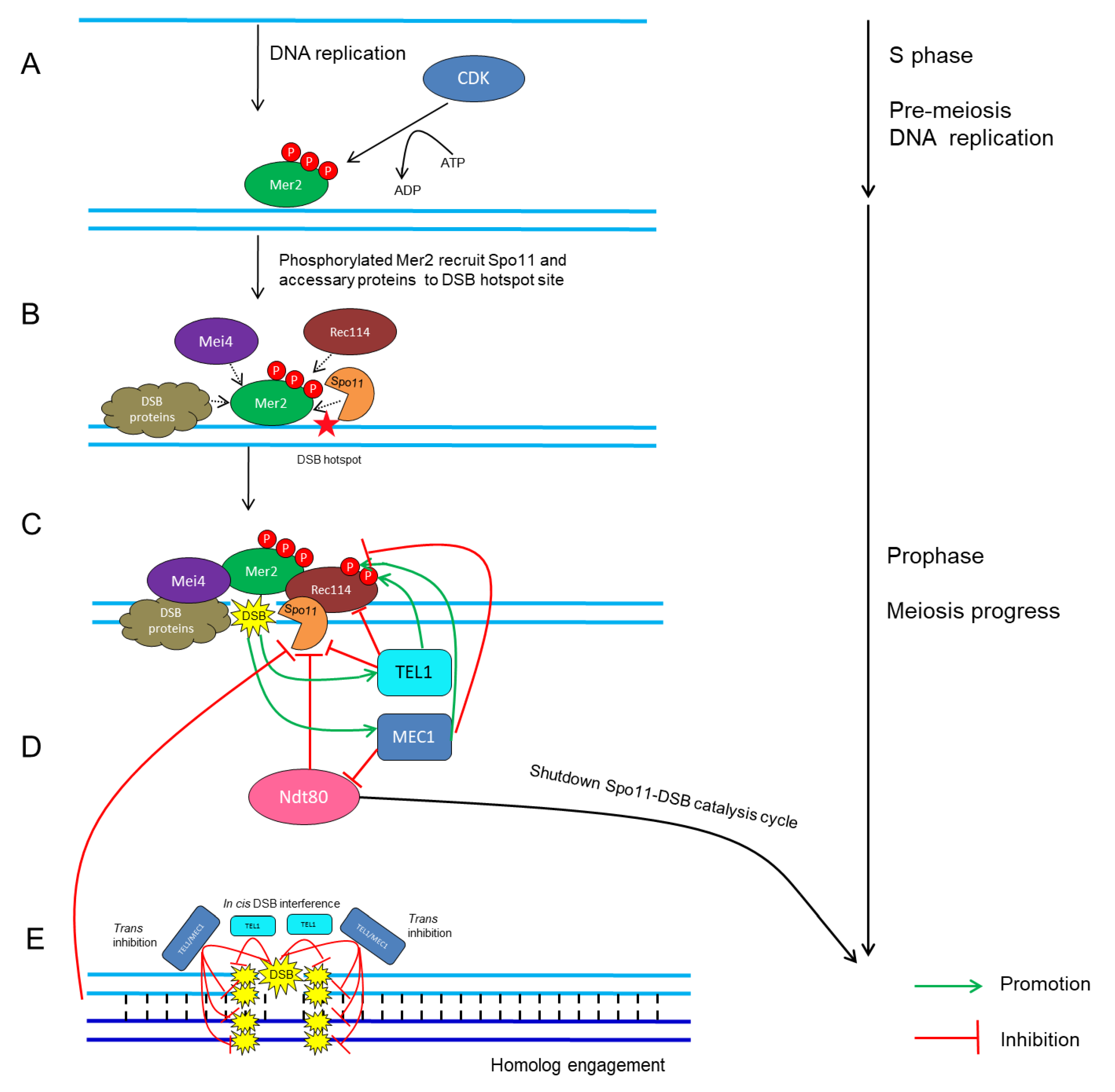
Figure 1. Schematic network of CDK- and ATM/ATR-mediated regulatory cycles of meiotic DSB timing and number in S. cerevisiae. (A) CDK phosphorylates Mer2 during pre-meiotic DNA replication [114,115,120][40][41][47]. (B) Phosphorylated Mer2 recruits Rec114, Mei4, Spo11 and other DSB proteins to DSB hotspot sites [45,120,121][46][47][48]. (C) DSB formation catalyzed by Spo11 and accessary proteins [4,66][4][49]. (D) Recurrent DSB formation activates TEL1/MEC1-dependent positive- and negative-feedback loops, which then restrains Spo11 activity and regulates the rate and number of DSB formation [136,140,147,156][50][51][52][53]. (E) Cis DSB interference mediated by TEL1 reduces the frequency of coincident DSB formation at the region adjacent to an already-formed DSB [143,149][54][55]. Trans inhibition mediated by TEL1 and MEC1 describes the ability of a DSB formation on one chromosome to suppress DSB formation on its homolog and sister chromatid at the same or adjacent regions [141][56].
3.2. Tel1/ATM and Mec1/ATR
Once DSBs are made, there are several mechanisms to maintain the proper number of DSBs within a chromosome region [95,111,136,137,138,139,140,141][33][50][51][56][72][73][74][75]. This phenomenon, termed DSB homeostasis, prevents the severe effects caused by excess or insufficient Spo11 activity [136,137,140,142,143][50][51][54][73][76]. In S. cerevisiae, Tel1 and the related protein kinase Mec1 act as key factors that synergistically fine-tune the number of DSBs [141,142,144][56][76][77]. Lack of Tel1 leads to a considerable increase in the level of Spo11-oligonucleotide complexes, which are the by-product of meiotic DSB formation, indicating that Tel1 negatively regulates DSB formation (Figure 1C) [95][72]. In contrast, a defect in Mec1 activity causes a dramatic reduction in DSB number via inhibition of Ndt80, which triggers the exit from pachytene and shuts down Spo11-DSB catalysis of the cell cycle [145,146][78][79]. Mec1 ensures persistent Spo11 activity and indirectly promotes DSB formation, revealing that Mec1 positively regulates DSB formation (Figure 1C) [147,148][52][80]. In addition, recent studies in S. cerevisiae have revealed a cis-regulatory machinery, termed DSB interference, that reduces the frequency of coincident DSBs at the region adjacent to the preexisting DSB [136[50][54][76],142,143], and this process is dependent on TEL1 over short distances of ~70–100 kb (Figure 1D) [95,149][55][72]. On the contrary, the other machinery, known as trans-inhibition, defines the ability of a DSB to occur on a single chromosome, and this suppresses DSB designation on its homolog and sister chromatid at the same locus [141][56]. This mechanism relies on both Tel1 and Mec1, ensuring that an interhomolog interaction surrounding a DSB will not occur twice at the allelic or two nearby chromosomal positions so that DSBs are constrained to one per pair of homologs (Figure 1E) [141][56]. Interestingly, homolog engagement can also restrict the number of DSBs by inhibiting Spo11 activity (Figure 1E) [101,150][27][81]. Moreover, Tel1 and/or Mec1 can phosphorylate Rec114 directly, which limits its interaction with DSB hotspots and consequently reduces DSB formation genome wide (Figure 1C) [140][51]. In Arabidopsis, Tel1 and Mec1 homologs exist, termed ATM and ATR, respectively. However, ATM and ATR play synergetic roles in maintenance of genomic stability in meiotic cells by processing Spo11-dependent DSBs rather than influencing DSB formation [151,152,153,154,155][82][83][84][85][86]. Therefore, either ATM/ATR-mediated signaling pathway is evolutionarily divergent across different species or the effects of ATM/ATR on DSB formation is too subtle to be detected by conventional methods.References
- Feng, X.; Zilberman, D.; Dickinson, H. A conversation across generations: Soma-germ cell crosstalk in plants. Dev. Cell 2013, 24, 215–225.
- Nakajima, K. Be my baby: Patterning toward plant germ cells. Curr. Opin. Plant Biol. 2018, 41, 110–115.
- Saito, T.T.; Colaiácovo, M.P. Regulation of Crossover Frequency and Distribution during Meiotic Recombination. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2018; Volume 82, pp. 223–234.
- Lam, I.; Keeney, S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014, 7, a016634.
- Mercier, R.; Mezard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015, 66, 297–327.
- Keeney, S.; Lange, J.; Mohibullah, N. Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu. Rev. Genet. 2014, 48, 187–214.
- Lambing, C.; Heckmann, S. Tackling Plant Meiosis: From Model Research to Crop Improvement. Front. Plant Sci. 2018, 9.
- Edlinger, B.; Schlogelhofer, P. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 2011, 62, 1545–1563.
- Wang, Y.; Copenhaver, G.P. Meiotic Recombination: Mixing It Up in Plants. Annu. Rev. Plant Biol. 2018.
- Zelkowski, M.; Olson, M.A.; Wang, M.; Pawlowski, W. Diversity and Determinants of Meiotic Recombination Landscapes. Trends Genet. TIG 2019.
- Hartung, F.; Wurz-Wildersinn, R.; Fuchs, J.; Schubert, I.; Suer, S.; Puchta, H. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 2007, 19, 3090–3099.
- De Muyt, A.; Vezon, D.; Gendrot, G.; Gallois, J.L.; Stevens, R.; Grelon, M. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 2007, 26, 4126–4137.
- Vrielynck, N.; Chambon, A.; Vezon, D.; Pereira, L.; Chelysheva, L.; De Muyt, A.; Mezard, C.; Mayer, C.; Grelon, M. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 2016, 351, 939–943.
- Waterworth, W.M.; Altun, C.; Armstrong, S.J.; Roberts, N.; Dean, P.J.; Young, K.; Weil, C.F.; Bray, C.M.; West, C.E. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007, 52, 41–52.
- Bleuyard, J.Y.; Gallego, M.E.; White, C.I. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 2004, 113, 197–203.
- Puizina, J.; Siroky, J.; Mokros, P.; Schweizer, D.; Riha, K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 2004, 16, 1968–1978.
- Ji, J.; Tang, D.; Wang, K.; Wang, M.; Che, L.; Li, M.; Cheng, Z. The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 2012, 72, 18–30.
- Wang, Y.; Jiang, L.; Zhang, T.; Jing, J.; He, Y. ZmCom1 Is Required for Both Mitotic and Meiotic Recombination in Maize. Front. Plant Sci. 2018, 9, 1005.
- Uanschou, C.; Siwiec, T.; Pedrosa-Harand, A.; Kerzendorfer, C.; Sanchez-Moran, E.; Novatchkova, M.; Akimcheva, S.; Woglar, A.; Klein, F.; Schlogelhofer, P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007, 26, 5061–5070.
- Sun, H.; Treco, D.; Szostak, J.W. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 1991, 64, 1155–1161.
- Zakharyevich, K.; Ma, Y.; Tang, S.; Hwang, P.Y.; Boiteux, S.; Hunter, N. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol. Cell 2010, 40, 1001–1015.
- Cao, L.; Alani, E.; Kleckner, N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 1990, 61, 1089–1101.
- Pan, J.; Sasaki, M.; Kniewel, R.; Murakami, H.; Blitzblau, H.G.; Tischfield, S.E.; Zhu, X.; Neale, M.J.; Jasin, M.; Socci, N.D.; et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 2011, 144, 719–731.
- Choi, K.; Henderson, I.R. Meiotic recombination hotspots—A comparative view. Plant J. 2015, 83, 52–61.
- Choi, K.; Zhao, X.; Tock, A.J.; Lambing, C.; Underwood, C.J.; Hardcastle, T.J.; Serra, H.; Kim, J.; Cho, H.S.; Kim, J.; et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 2018, 28, 532–546.
- Lange, J.; Yamada, S.; Tischfield, S.E.; Pan, J.; Kim, S.; Zhu, X.; Socci, N.D.; Jasin, M.; Keeney, S. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell 2016, 167, 695–708.
- Thacker, D.; Mohibullah, N.; Zhu, X.; Keeney, S. Homologue engagement controls meiotic DNA break number and distribution. Nature 2014, 510, 241–246.
- Underwood, C.J.; Choi, K.; Lambing, C.; Zhao, X.; Serra, H.; Borges, F.; Simorowski, J.; Ernst, E.; Jacob, Y.; Henderson, I.R.; et al. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 2018, 28, 519–531.
- Smagulova, F.; Gregoretti, I.V.; Brick, K.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 2011, 472, 375–378.
- Brick, K.; Smagulova, F.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genetic recombination is directed away from functional genomic elements in mice. Nature 2012, 485, 642–645.
- He, Y.; Wang, M.; Dukowic-Schulze, S.; Zhou, A.; Tiang, C.L.; Shilo, S.; Sidhu, G.K.; Eichten, S.; Bradbury, P.; Springer, N.M.; et al. Genomic features shaping the landscape of meiotic double-strand-break hotspots in maize. Proc. Natl. Acad. Sci. USA 2017, 114, 12231–12236.
- Zhu, X.; Keeney, S. High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae. Genetics 2015, 201, 525–542.
- Mohibullah, N.; Keeney, S. Numerical and spatial patterning of yeast meiotic DNA breaks by Tel1. Genome Res. 2017, 27, 278–288.
- Lam, I.; Keeney, S. Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science 2015, 350, 932–937.
- Fowler, K.R.; Sasaki, M.; Milman, N.; Keeney, S.; Smith, G.R. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014, 24, 1650–1664.
- Tock, A.J.; Henderson, I.R. Hotspots for Initiation of Meiotic Recombination. Front. Genet. 2018, 9, 521.
- Yamada, S.; Kim, S.; Tischfield, S.E.; Jasin, M.; Lange, J.; Keeney, S. Genomic and chromatin features shaping meiotic double-strand break formation and repair in mice. Cell Cycle 2017, 16, 1870–1884.
- Vader, G.; Blitzblau, H.G.; Tame, M.A.; Falk, J.E.; Curtin, L.; Hochwagen, A. Protection of repetitive DNA borders from self-induced meiotic instability. Nature 2011, 477, 115–119.
- Sasaki, M.; Tischfield, S.E.; van Overbeek, M.; Keeney, S. Meiotic Recombination Initiation in and around Retrotransposable Elements in Saccharomyces cerevisiae. PLoS Genet. 2013, 9, e1003732.
- Bustamante-Jaramillo, L.F.; Ramos, C.; Alonso, L.; Sesmero, A.; Segurado, M.; Martin-Castellanos, C. CDK contribution to DSB formation and recombination in fission yeast meiosis. PLoS Genet. 2019, 15, e1007876.
- Wan, L.; Niu, H.; Futcher, B.; Zhang, C.; Shokat, K.M.; Boulton, S.J.; Hollingsworth, N.M. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008, 22, 386–397.
- Benjamin, K.R. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003, 17, 1524–1539.
- Nasmyth, K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr. Opin. Cell Biol. 1993, 5, 166–179.
- Stuart, D.; Wittenberg, C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998, 12, 2698–2710.
- Smith, K.N.; Penkner, A.; Ohta, K.; Klein, F.; Nicolas, A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 2001, 11, 88–97.
- Li, J.; Hooker, G.W.; Roeder, G.S. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 Form a Complex Required for Meiotic Double-Strand Break Formation. Genetics 2006, 173, 1969.
- Sasanuma, H.; Hirota, K.; Fukuda, T.; Kakusho, N.; Kugou, K.; Kawasaki, Y.; Shibata, T.; Masai, H.; Ohta, K. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 2008, 22, 398–410.
- Keeney, S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001, 52, 1–53.
- Panizza, S.; Mendoza, M.A.; Berlinger, M.; Huang, L.; Nicolas, A.; Shirahige, K.; Klein, F. Spo11-Accessory Proteins Link Double-Strand Break Sites to the Chromosome Axis in Early Meiotic Recombination. Cell 2011, 146, 372–383.
- Lukaszewicz, A.; Lange, J.; Keeney, S.; Jasin, M. Control of meiotic double-strand-break formation by ATM: Local and global views. Cell Cycle 2018, 17, 1155–1172.
- Carballo, J.A.; Panizza, S.; Serrentino, M.E.; Johnson, A.L.; Geymonat, M.; Borde, V.; Klein, F.; Cha, R.S. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet. 2013, 9, e1003545.
- Gray, S.; Allison, R.M.; Garcia, V.; Goldman, A.S.; Neale, M.J. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol. 2013, 3, 130019.
- Prugar, E.; Burnett, C.; Chen, X.; Hollingsworth, N.M. Coordination of Double Strand Break Repair and Meiotic Progression in Yeast by a Mek1-Ndt80 Negative Feedback Loop. Genetics 2017, 206, 497–512.
- Xu, L.; Kleckner, N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995, 14, 5115–5128.
- Cooper, T.J.; Garcia, V.; Neale, M.J. Meiotic DSB patterning: A multifaceted process. Cell Cycle 2016, 15, 13–21.
- Zhang, L.; Kim, K.P.; Kleckner, N.E.; Storlazzi, A. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. USA 2011, 108, 20036–20041.
- Boudolf, V.; Barrôco, R.; de Almeida Engler, J.; Verkest, A.; Beeckman, T.; Naudts, M.; Inzé, D.; De Veylder, L. B1-Type Cyclin-Dependent Kinases Are Essential for the Formation of Stomatal Complexes in Arabidopsis thaliana. Plant Cell 2004, 16, 945–955.
- Boniotti, M.B.; Gutierrez, C. A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 2001, 28, 341–350.
- Wijnker, E.; Harashima, H.; Müller, K.; Parra-Nuñez, P.; de Snoo, C.B.; van de Belt, J.; Dissmeyer, N.; Bayer, M.; Pradillo, M.; Schnittger, A. The Cdk1/Cdk2 homolog CDKA;1 controls the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 12534–12539.
- Corellou, F.; Camasses, A.; Ligat, L.; Peaucellier, G.; Bouget, F.Y. Atypical regulation of a green lineage-specific B-type cyclin-dependent kinase. Plant Physiol. 2005, 138, 1627–1636.
- Adachi, S.; Uchimiya, H.; Umeda, M. Expression of B2-type cyclin-dependent kinase is controlled by protein degradation in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 1683–1686.
- Bulankova, P.; Akimcheva, S.; Fellner, N.; Riha, K. Identification of Arabidopsis Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes. PLoS Genet. 2013, 9, e1003508.
- Potuschak, T.; Doerner, P. Cell cycle controls: Genome-wide analysis in Arabidopsis. Curr. Opin. Plant Biol. 2001, 4, 501–506.
- Nowack, M.K.; Harashima, H.; Dissmeyer, N.; Zhao, X.A.; Bouyer, D.; Weimer, A.K.; De Winter, F.; Yang, F.; Schnittger, A. Genetic Framework of Cyclin-Dependent Kinase Function in Arabidopsis. Dev. Cell 2012, 22, 1030–1040.
- Richard, C.; Granier, C.; Inze, D.; De Veylder, L. Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J. Exp. Bot. 2001, 52, 1625–1633.
- Wijnker, E.; Schnittger, A. Control of the meiotic cell division program in plants. Plant Reprod. 2013, 26, 143–158.
- Zheng, T.; Nibau, C.; Phillips, D.W.; Jenkins, G.; Armstrong, S.J.; Doonan, J.H. CDKG1 protein kinase is essential for synapsis and male meiosis at high ambient temperature in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 2182–2187.
- Wu, Z.; Ji, J.; Tang, D.; Wang, H.; Shen, Y.; Shi, W.; Li, Y.; Tan, X.; Cheng, Z.; Luo, Q. OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 2015, 6, 21.
- Azumi, Y.; Liu, D.; Zhao, D.; Li, W.; Wang, G.; Hu, Y.; Ma, H. Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 2002, 21, 3081–3095.
- D’Erfurth, I.; Cromer, L.; Jolivet, S.; Girard, C.; Horlow, C.; Sun, Y.; To, J.P.; Berchowitz, L.E.; Copenhaver, G.P.; Mercier, R. The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 2010, 6, e1000989.
- Bulankova, P.; Riehs-Kearnan, N.; Nowack, M.K.; Schnittger, A.; Riha, K. Meiotic progression in Arabidopsis is governed by complex regulatory interactions between SMG7, TDM1, and the meiosis I-specific cyclin TAM. Plant Cell 2010, 22, 3791–3803.
- Garcia, V.; Gray, S.; Allison, R.M.; Cooper, T.J.; Neale, M.J. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 2015, 520, 114–118.
- Cooper, T.J.; Wardell, K.; Garcia, V.; Neale, M.J. Homeostatic regulation of meiotic DSB formation by ATM/ATR. Exp. Cell Res. 2014, 329, 124–131.
- Penedos, A.; Johnson, A.L.; Strong, E.; Goldman, A.S.; Carballo, J.A.; Cha, R.S. Essential and Checkpoint Functions of Budding Yeast ATM and ATR during Meiotic Prophase Are Facilitated by Differential Phosphorylation of a Meiotic Adaptor Protein, Hop1. PLoS ONE 2015, 10, e0134297.
- Shiloh, Y. The ATM-mediated DNA-damage response: Taking shape. Trends Biochem. Sci. 2006, 31, 402–410.
- Lange, J.; Pan, J.; Cole, F.; Thelen, M.P.; Jasin, M.; Keeney, S. ATM controls meiotic double-strand-break formation. Nature 2011, 479, 237–240.
- Blitzblau, H.G.; Hochwagen, A. ATR/Mec1 prevents lethal meiotic recombination initiation on partially replicated chromosomes in budding yeast. eLife 2013, 2, e00844.
- Xu, L.; Ajimura, M.; Padmore, R.; Klein, C.; Kleckner, N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 6572.
- Winter, E. The Sum1/Ndt80 Transcriptional Switch and Commitment to Meiosis in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2012, 76, 1.
- Argunhan, B.; Farmer, S.; Leung, W.K.; Terentyev, Y.; Humphryes, N.; Tsubouchi, T.; Toyoizumi, H.; Tsubouchi, H. Direct and indirect control of the initiation of meiotic recombination by DNA damage checkpoint mechanisms in budding yeast. PLoS ONE 2013, 8, e65875.
- Kauppi, L.; Barchi, M.; Lange, J.; Baudat, F.; Jasin, M.; Keeney, S. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013, 27, 873–886.
- Kim, J.H.; Ryu, T.H.; Lee, S.S.; Lee, S.; Chung, B.Y. Ionizing radiation manifesting DNA damage response in plants: An overview of DNA damage signaling and repair mechanisms in plants. Plant Sci. 2019, 278, 44–53.
- Culligan, K.M.; Britt, A.B. Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 2008, 55, 629–638.
- Garcia, V.; Bruchet, H.; Camescasse, D.; Granier, F.; Bouchez, D.; Tissier, A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003, 15, 119–132.
- Eckardt, N.A. ATM to the rescue: Repairing DNA damage. Plant Cell 2003, 15, 1–3.
- Roitinger, E.; Hofer, M.; Kocher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlogelhofer, P.; Mechtler, K. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteom. MCP 2015, 14, 556–571.
More
